Abstract
Corynespora leaf spot caused by Corynespora cassiicola is one of the major diseases in cucumber (Cucumis sativus L.). However, the resistance mechanisms and signals of cucumber to C. cassiicola are unclear. Here, we report that the mildew resistance locus O (MLO) genes, CsMLO1 and CsMLO2, are both negative modulators of the cucumber defense response to C. cassiicola. Subcellular localization analysis showed that CsMLO1 and CsMLO2 are localized in the plasma membrane. Expression analysis indicated that the transcript levels of CsMLO1 and CsMLO2 are linked to the defense response to C. cassiicola. Transient overexpression of either CsMLO1 or CsMLO2 in cucumber cotyledons reduced resistance to C. cassiicola, whereas silencing of either CsMLO1 or CsMLO2 enhanced resistance to C. cassiicola. The relationships of pathogenesis-related proteins, reactive oxygen species (ROS)-associated genes, and abscisic acid (ABA)-related genes to the overexpression and silencing of CsMLO1/CsMLO2 in non-infested cucumber plants were investigated. The results indicated that CsMLO1 mediated resistance against C. cassiicola by regulating the expression of pathogenesis-related proteins and ROS-associated genes, as well as through ABA signaling pathway-associated genes. The CsMLO2-mediated resistance against C. cassiicola primarily involves regulation of the expression of pathogenesis-related proteins. Our findings will guide strategies to enhance the resistance of cucumber to corynespora leaf spot.
1. Introduction
Cucumber target leaf spot (TLS) is caused by Corynespora cassiicola (Berk and Curt, Wei) and has resulted in increasing losses in cucumber production in recent years. Thus far, research on TLS has primarily focused on identification, control methods and biological characteristics. Several resistant varieties, including Jinyou 3 [1], Jinyou38 [1], Marketmore97 [2], HygBrid72502 [3], and USDA6623E [4], have been identified by disease resistance phenotypes in both Asia and Europe. At present, the inheritance of TLS resistance in cucumber has been found in several studies by using marker-assisted selection (MAS) and simple sequence repeat (SSR) techniques. A single dominant gene, Cca, show TLS resistance in the Royal Sluis 72502 cucumber line [5], while the single recessive genes cca-1, cca-2 and cca-3 show resistance to cucumber TLS [1,6,7]. Therefore, the controversy about the genetic control of TLS resistance, and whether TLS resistance is controlled by a single dominant or recessive gene remain unclear. In addition, the genetic and physical distances of linked markers from these studies are still far away from the target gene. These markers are not breeder-friendly and not amendable for high-throughput genotyping. Most plants have non-host resistance to microbial invaders, and this resistance mechanism provides broad-spectrum and strong resistance to non-adapted pathogens in plants [8]. The mildew resistance locus O (MLO) gene in non-host defense plays an important role in the model system of the interaction of barley with Blumeria graminis f. sp. hordei (Bgh) [9,10]. Inducible defense responses in nonhost plants is mainly caused by the hypersensitive response (HR) of host plants, including the accumulation of ROS, the activation of pathogenesis-related genes, and localized reinforcement of the plant cell walls [11,12]. Therefore, we can screen related resistance genes and study their molecular mechanisms of host resistance to cucumber TLS. Elucidating the molecular mechanism of cucumber-C. cassiicola interactions is important for establishing ideal varieties.
MLO genes are considered negative regulators of plant immunity [13]. For instance, Arabidopsis MLO2, MLO6 and MLO12 have genetic redundancy in which Atmlo2 Atmlo6 Atmlo12 triple mutants are fully resistant to the powdery mildew pathogen and Atmlo2 single mutants are partially resistant [14,15,16]. Subsequent studies revealed that leaf cells show spontaneous cell death, callose deposition and reactive oxygen species (ROS) accumulation in the genetic background of Atmlo2 Atmlo6 Atmlo12 triple mutants and Atmlo2 single mutants [17]. In addition, after inoculation with the bacterial pathogen Pseudomonas syringae, induction of Arabidopsis MLO2 gene expression is associated with salicylic acid (SA) signaling, and systemically enhanced in plant leaves exhibiting systemic acquired resistance (SAR) [18]. Furthermore, it may be worth mentioning that Arabidopsis mlo2 mutants are defective in SAR against P. syringae [18]. At present, MLO genes have been implicated in other resistance pathways. Barley mlo alleles enhance susceptibility to the necrosis-inducing toxin from Bipolaris sorokiniana [19]. Studies have identified the interactions between Arabidopsis and bacteria (e.g., Pseudomonas syringae), Arabidopsis and oomycetes (e.g., Hyaloperonospora arabidopsidis and Albugo spp.), and Arabidopsis and fungus (Colletotrichum higginsianum) [20,21,22,23]. Furthermore, research has recently shown that the triple mutant line mlo2-5 mlo6-2 mlo12-1 increased susceptibility to P. syringae pv. Maculicola [24]. In our laboratory, transcriptome and iTRAQ analyses identified two CsMLOs of cucumber that are involved in the response to C. cassiicola [25,26]. However, the CsMLOs function in C. cassiicola infection, and the defense mechanisms have not been described in cucumber.
Plant self-resistance causes a series of intracellular signaling responses under stress, and they include physiological and biochemical processes. ROS are important signal transduction molecules involved in different cellular signaling pathways [27,28]. The main sources of ROS production include superoxide radicals (O2·−), hydroxyl radicals (OH), hydrogen peroxide (H2O2) and singlet oxygen [29,30]. In plant immunity, ROS production in response to pathogen recognition is mediated by the In plant immunity, ROS production in response to pathogen recognition is mediated by the oxidase nicotinamide adenine dinucleotide phosphate (NADPH) oxidases, also called respiratory burst oxidase homologs (RBOHs), and arabidopsis AtRBOHD and AtRBOHF plays a key role in apoplastic ROS generation during plant-pathogen interactions [31,32]. Moreover, the outbreak of ROS, cell death that is phenotypically similar to a hypersensitive response (HR) and increased levels of ROS-producing enzymes play an active role in plant defense [33,34]. Additionally, stoma closure participates in H2O2 production in Arabidopsis, which is promoted in guard cells by pathogen-associated molecular patterns (PAMPs)-induced abscisic acid (ABA) signaling, and this pattern increases plant resistance to pathogens [35,36,37]. Therefore, ABA is involved in the development of defense responses during plant-pathogen interactions to block pathogen invasion. Three core components include the ABA receptors PYLs, the negative regulator type 2C protein phosphatases (PP2Cs) and positive regulator subfamily 2 of SNF1-related kinases (SnRK2s) in ABA signal transduction plants [38]. ABA-insensitive 5 (ABI5), as a basic leucine-zipper (bZIP) transcription factor, is essential to ABA-mediated developmental checkpoints [39]. In cucumber seedlings, CsPYL2, CsPP2C2 and CsSnKR2.2 respond to the ABA signaling pathway under drought stress [38]. Furthermore, pathogenesis-related proteins (PR proteins) proteins, including pathogenesis-related protein 1-1a (PR1-1a), β-1, 3-glucanase (PR2) and chitinase (PR3), are increased to promote resistance to pathogens [40,41]. However, these aspects of ROS and ABA signal transduction remain largely unexplored in CsMLOs-C. cassiicola interactions.
In this study, CsMLO1 (XM_004148737) and CsMLO2 (XM_004142345) were shown to be induced by C. cassiicola. A subcellular localization analysis showed that these proteins are located on the plasma membrane. CsMLO1 and CsMLO2 respond differently to abiotic stresses. An investigation of their defense roles via transient CsMLO1/CsMLO2-silencing and CsMLO1/CsMLO2-overexpressing cucumber plants showed that CsMLO1 and CsMLO2 both function as negative modulators in the host resistance response to C. cassiicola. Furthermore, CsMLO1 silencing might participate in the defense response to C. cassiicola by inducing the significant upregulation of several ROS-associated genes, ABA signaling pathway-associated genes or pathogenesis-related proteins. We concluded that CsMLO1 participated in the defense response to C. cassiicola in plants through multiple signaling regulatory networks. CsMLO2 regulated the expression of pathogenesis-related proteins in uninfected cucumbers. There was no pattern in the expression of ROS- and ABA-related genes in CsMLO2 transgenic cucumbers, suggesting that this gene might regulate other signaling pathways that affect cucumber resistance to C. cassiicola.
2. Results
2.1. Phenotypic Characterization of Cucumber Cultivars after C. cassiicola Infection
All cucumber cultivars were inoculated on the second leaves with C. cassiicola and infected after 5 days. The disease index statistics of each strain are shown in Table 1, which shows that there were obvious differences in disease resistance among different cucumber strains. Jinyou 38 was highly resistant to C. cassiicola. B21-a-2-1-2 and F10 were moderately resistant to C. cassiicola. B21-a-2-2-2, 995, Jinyan 4 and Xintaimici were susceptible to C. cassiicola. Based on the disease index survey, we selected the high-resistance variety Jinyou 38 and the susceptible variety Xintaimici to carry out the next experiments [42].

Table 1.
The disease index of different cucumber cultivars to C.cassiicola.
2.2. Isolation and Phylogenetic Analysis of CsMLO1 and CsMLO2
A search of the cucumber genomics database (http://cucurbitgenomics.org/) and gene prediction analysis of the cucumber genome led to the identification of two hypothetical full-length CsMLO1 and CsMLO2 transcripts in the Xintaimici and Jinyou 38 cultivars. The 1749 bp full-length CsMLO1 cDNA encoding a 66.92 kDa protein with 582 amino acid residues and the 1725 bp full-length CsMLO2 cDNA encoding a 64.25 kDa protein with 574 amino acid residues were identified from cucumber leaves (Figures S1 and S2). The cDNA sequences of CsMLO1 were cloned from the experimental materials of Jinyou 38 and Xintaimici, and the results showed that there was no difference in cDNA sequences of CsMLO1 between the Jinyou 38 variety and Xintaimici variety. Similarly, similar cloning and alignment methods found that cDNA sequences of CsMLO2 also had no differences between the Jinyou 38 variety and Xintaimici variety. A sequence analysis predicted that CsMLO1 and CsMLO2 were homologous to MLO proteins. Both homoeologous proteins shared high similarity with proteins for powdery mildew fungus susceptibility in pepper, Arabidopsis, turnip and barley. These amino acid sequences were highly conserved and contained seven transmembrane structures (TM1–TM7) and a C-terminal calmodulin-binding domain (CaMBD) in the C-terminal region (Figure S3).
A phylogenetic tree containing 21 MLO proteins from dicots and monocots demonstrated the phylogenetic relationships. CsMLO1 and CsMLO2 proteins were clustered with the five dicot subfamilies containing AtMLO2, AtMLO6, AtMLO12, BrMLO1, LeMLO1, and CaMLO1 and the monocot OsMLO1 (Figure S4A). Each of these proteins was previously shown to be required for powdery mildew susceptibility. Putative CaMBDs in the C-terminus of CsMLO1 and CsMLO2 were located at Ala 453–Arg 471 and Ala 443–Arg 461 (Figure S4B), which was highly conserved throughout the Arabidopsis MLO family. The Ca2+-dependent CaM-binding motif contained Trp 419, which is known to play a significant role in CaM binding to CaM target proteins [43]. The potential functional conservation of CsMLO1 and CsMLO2 suggested that they were co-orthologs of the monocot barley HvMLO and the dicot Arabidopsis AtMLO2, AtMLO6, and AtMLO12 proteins.
2.3. Subcellular Localization and Expression Patterns of CsMLO1 and CsMLO2
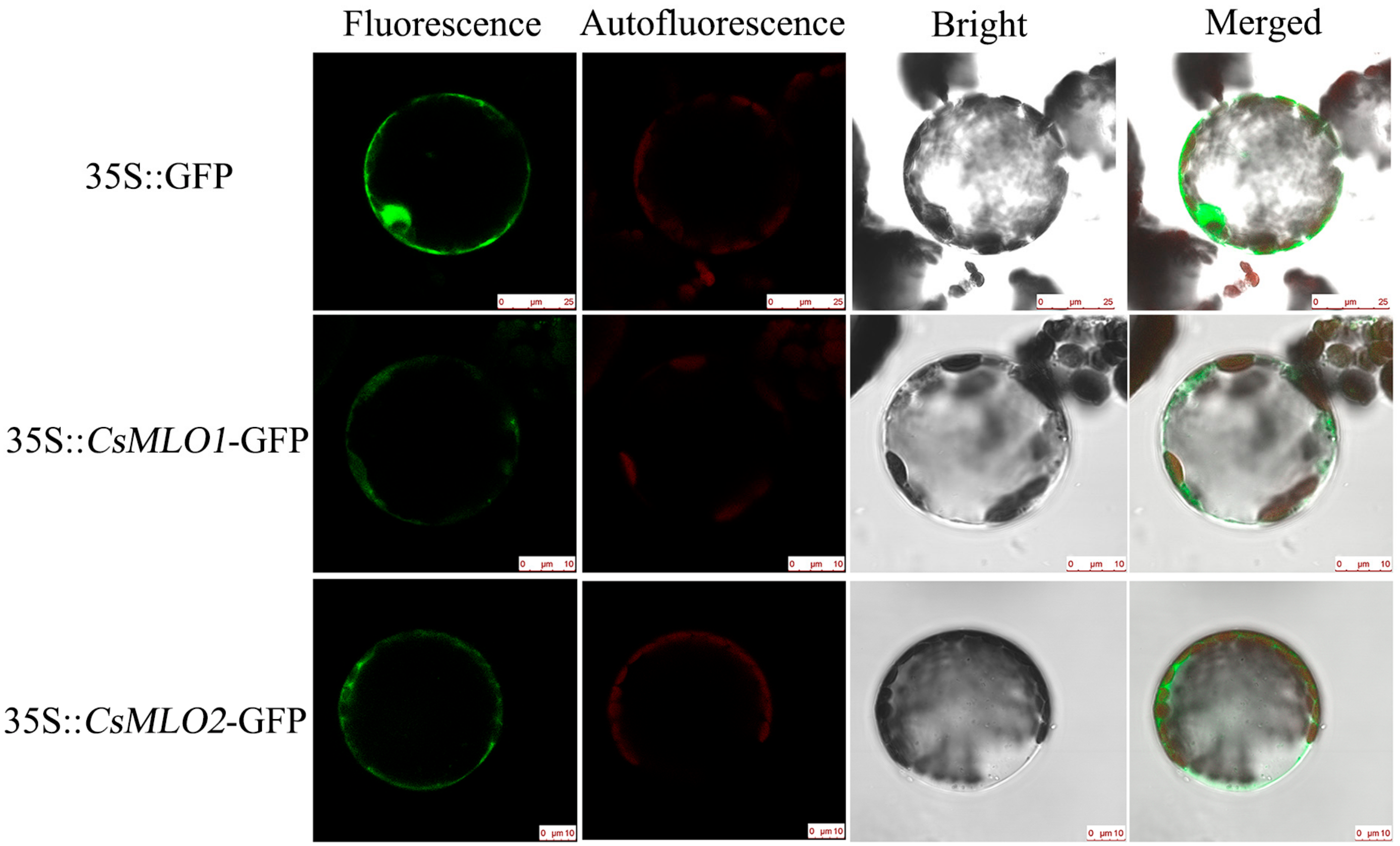
A sequence analysis indicated that the proteins encoded by CsMLO1 and CsMLO2 were transmembrane proteins with seven transmembrane structures (Figure S5). To confirm this prediction, we determined the subcellular localizations of the CsMLO1 and CsMLO2 proteins, and confocal imaging showed that CsMLO1-GFP and CsMLO2-GFP fluorescence were localized in the plasma membrane, whereas the GFP protein was found in multiple subcellular compartments, including the cell membrane, cytoplasm, and nucleus (Figure 1).
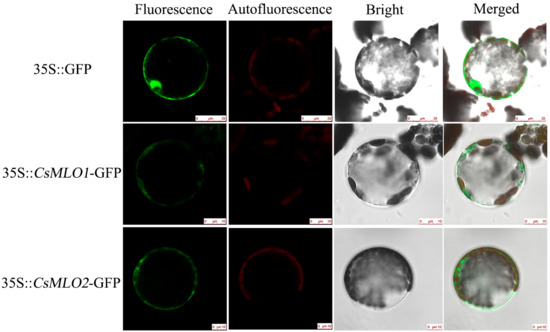
Figure 1.
Subcellular localization of CsMLO1 and CsMLO2. Green fluorescence protein (GFP) was detected in the N. benthamiana protoplasts of the 35S::GFP, 35S::CsMLO1-GFP and 35S::CsMLO2-GFP constructs. CsMLO1::GFP and CsMLO2::GFP localized in the plasma membrane, GFP protein alone localized throughout the whole cells. Fluorescence, chloroplast autofluorescence, bright field and merged images were obtained using a Leica confocal microscope. Scale bars = 10 μm.
To investigate the spatial expression patterns of CsMLO1 and CsMLO2, we performed a qRT-PCR analysis of the tissue-specific expression patterns in different cucumber organs of susceptible and resistant cultivars (Figure S6). CsMLO1 exhibited the highest expression in the stem followed by the leaf, but it was weakly expressed in the cotyledon and root from the susceptible cultivar (Figure S6A). However, in the resistant cultivars, CsMLO1 was relatively highly expressed in the cotyledons and roots, followed by the leaves and stems (Figure S6B). In addition, CsMLO2 was significantly expressed in the leaf and cotyledon but was barely expressed in the stem and root from different cucumber organs of susceptible and resistant cultivars (Figure S6C,D). Therefore, the combined subcellular localization and spatial expression patterns indicated that CsMLO1 and CsMLO2 are both typical MLO proteins and exhibit tissue-specific expression.
2.4. Response to Abiotic Stresses
We investigated the expression patterns of CsMLO1 and CsMLO2 genes in response to plant hormones and abiotic stimuli. The transcript levels of CsMLO1 and CsMLO2 were observed under abiotic stimuli in the leaves of susceptible and resistant cultivars. To determine whether CsMLO1 and CsMLO2 were regulated by extracellular signals, we investigated the possible involvement of CsMLO1 and CsMLO2 in signaling pathways related to phytohormones and CaCl2. The expression characteristics of the CsMLO1 gene and the CsMLO2 gene were distinct in the resistant cultivar (Figure S7). Compared with those of the control plants, CsMLO1 and CsMLO2 showed different expression patterns after exogenous treatment with H2O2, ABA, SA, MeJA and CaCl2. The CsMLO1 transcript level was suppressed in resistant cultivars after treatment with exogenous ABA, CaCl2 and H2O2. CsMLO2 transcription was downregulated at 24 h and 48 h after treatment with exogenous H2O2, and CsMLO2 expression was distinctly induced and peaked at 12 h, followed by a downward trend in expression at 24 h and 48 h after treatment with exogenous CaCl2. But, CsMLO2 expression was upregulated after treatment with exogenous SA. Thus, these results indicated that CsMLO1 and CsMLO2 might be involved in ROS, ABA, SA and calcium signaling pathway.
2.5. Expression Patterns of CsMLO1 and CsMLO2 in the Resistant/Susceptible Cucumber Cultivars after C. cassiicola Infection
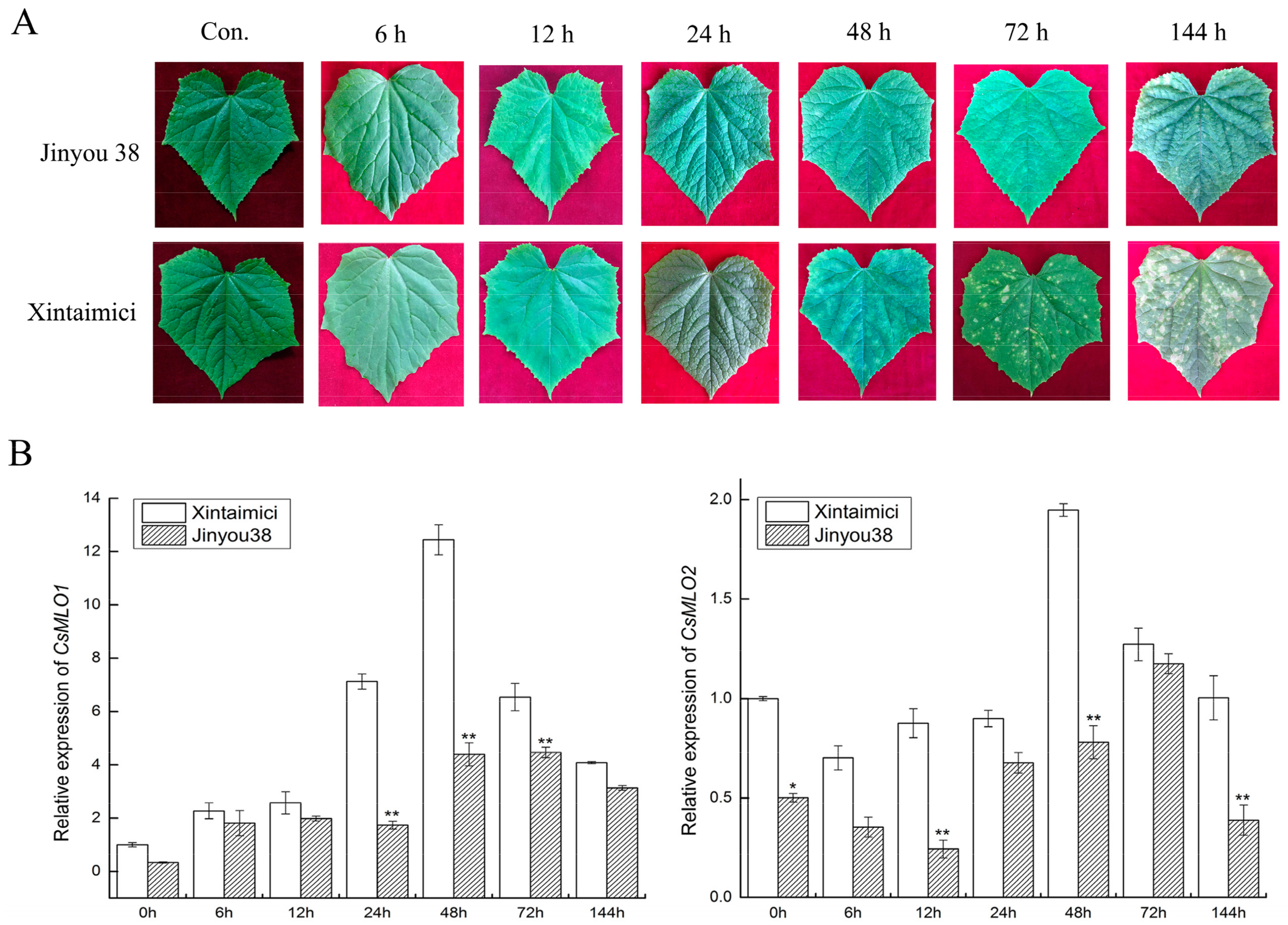
CsMLO1 and CsMLO2 of cucumber were found to be involved in the response to C. cassiicola in previous work by iTRAQ and RNA-seq, and resistant/susceptible cucumber cultivars were also investigated for disease index by disease index analysis (Table S1). Jinyou 38 showed resistance with a low infection index (14.85), and Xintaimici showed high susceptibility with a 72.33 index at 7 dpi (days post-inoculation). To further confirm these results, we obtained images of the cucumber cultivars in response to C. cassiicola under different time courses. Susceptible cucumber plants showed symptoms at 48 h after C. cassiicola infection, and evident symptoms were found on the leaves at 72 h and 144 h, whereas only faint symptoms were detected on the leaves of the resistant cucumber plants at 72 h and 144 h (Figure 2A). To determine whether CsMLO1 and CsMLO2 were affected by C. cassiicola, we performed RT-qPCR assays to examine the expression patterns of CsMLO1 and CsMLO2 at different times using resistant and susceptible cultivars (Figure 2B). A comparison of the two cultivars showed that CsMLO1 expression was induced at 24 h and peaked at 48 h in the susceptible cultivar. In the resistant cultivar, CsMLO1 expression was largely induced at 48 and 72 h. However, CsMLO2 expression was decreased in the initial phase in the two cultivars and appeared to be significantly higher in the susceptible cultivar than in the resistant cultivar at 48 h. This finding suggested that the expression of CsMLO1 and CsMLO2 appeared to be faster in the susceptible cultivar than in the resistant cultivar when inoculated with C. cassiicola.
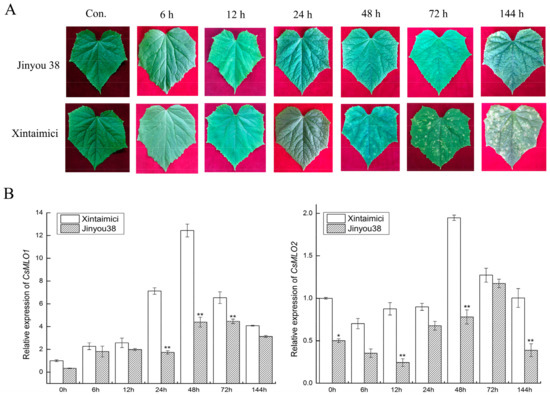
Figure 2.
The phenotype and expression patterns of CsMLO in Jinyou38 and Xintaimici cultivars were inoculated with C. cassiicola. (A) Symptoms analysis in leaves of Jinyou 38 and Xintaimici cultivars after C. cassiicola challenge. (B) The transcript levels of CsMLO1 and CsMLO2 are shown in Jinyou 38 and Xintaimici cultivars after inoculation of C. cassiicola. Expression analysis of candidate genes at 0, 6, 12, 24, 48, 72 and 144 hpi (hours post-inoculation) using the 2−ΔΔCt method. Data are means (± standard deviation (SD)) of three biological replicates per cultivars. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01). Jinyou38 = high-resistant cultivar. Xintaimici = susceptible cultivar.
2.6. CsMLO1 or CsMLO2 Silencing Improves Resistance to C. cassiicola in Cucumber Cotyledon
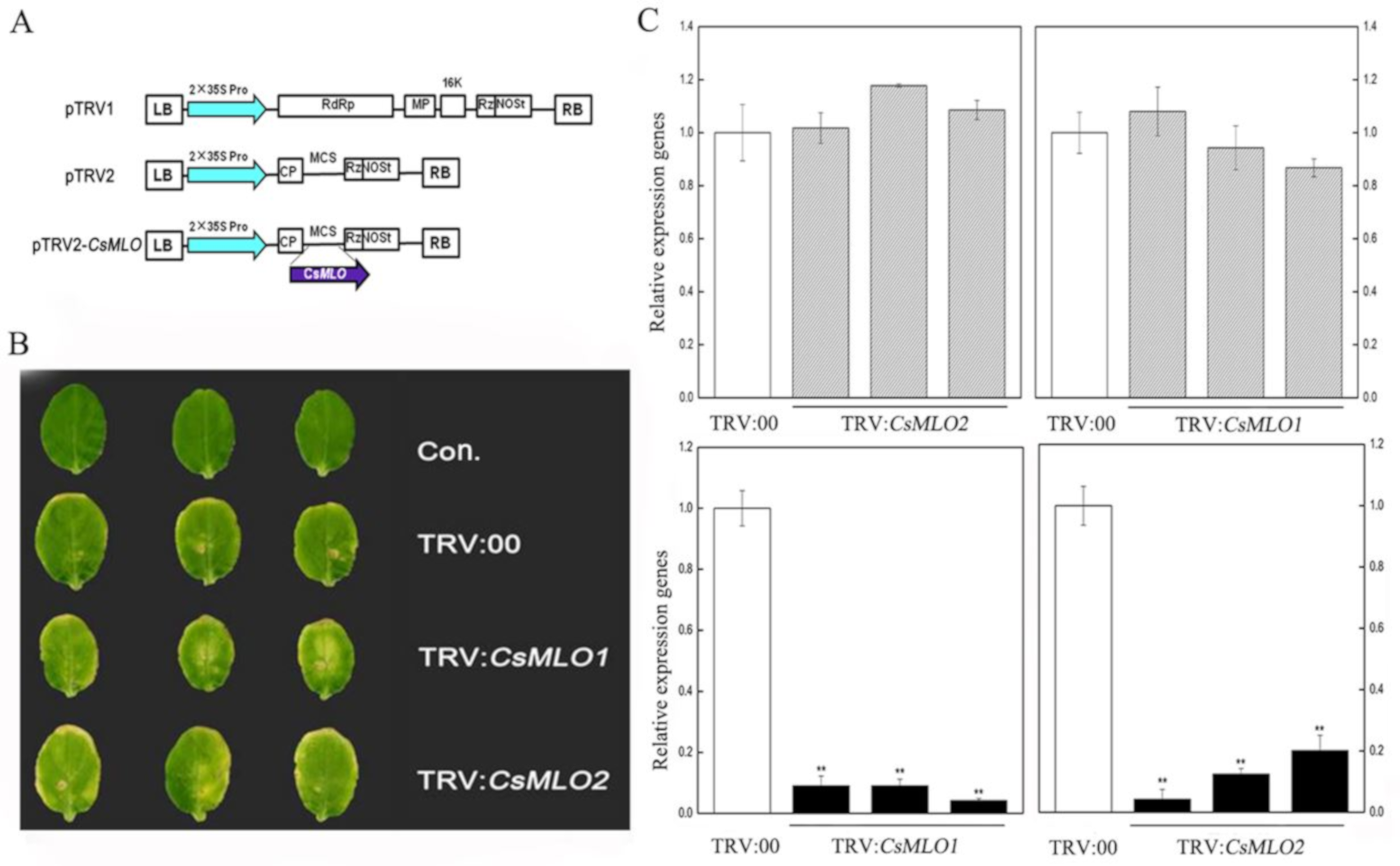
To determine whether a loss-of-function of CsMLO1 and CsMLO2 affected cucumber resistance to C. cassiicola, we used tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) to silence CsMLO1/CsMLO2 transcripts in the susceptible cultivar Xintaimici. A 3′-terminal fragment specific to CsMLO1/CsMLO2 was inserted in the vector pTRV2, the pTRV2-CsMLO1/CsMLO2 recombinant plasmids (Figure 3A). Subsequently, TRV:CsMLO1 or TRV:CsMLO2 virus was injected in cucumber cotyledons. The chlorotic mosaic symptoms of TRV:00, TRV:CsMLO1, TRV:CsMLO2 emerged in the cotyledons of transgenic cucumber plants, while no symptoms appeared in the non-injected cucumber cotyledons, suggesting that TRV successfully invaded the cucumber plants (Figure 3B). Simultaneously, efficient silencing of CsMLO1 and CsMLO2 was confirmed by RT-qPCR (Figure 3C). In addition, to test whether the CsMLO2 gene was also silenced in CsMLO1-silenced cucumber plants, we detected CsMLO2 expression and observed no silencing by RT-qPCR. Similarly, we detected CsMLO1 expression in CsMLO2-silenced cucumber cotyledon. These results indicated that expression was almost abolished in CsMLO1-silenced and CsMLO2-silenced cucumbers.
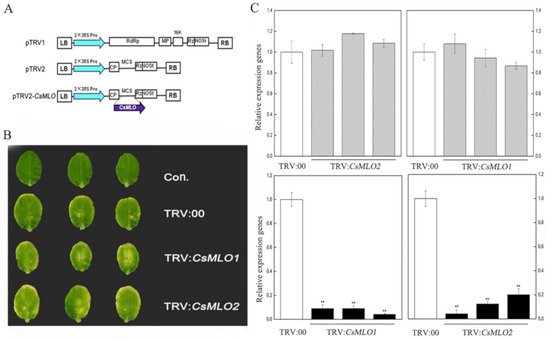
Figure 3.
Identification of CsMLO-silencing cucumber plants. (A) Schematic of the CsMLO1-silenced and CsMLO2-silenced constructs. (B) Symptoms in detached cotyledons of silencing cucumber plants. Con. indicates non-injected plants. (C) CsMLO1-silenced and CsMLO2-silenced were identified in transgenic plants by RT-qPCR. Black bars showed the efficient silencing of CsMLO1 and CsMLO2 by RT-qPCR, and gray bars represented efficient silencing of CsMLO2 in CsMLO1-silenced cucumber cotyledons and efficient silencing of CsMLO1 in CsMLO2-silenced cucumber cotyledons. Data are means ± standard deviations from three independent experiments, and each column represents a sample containing three of cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01).
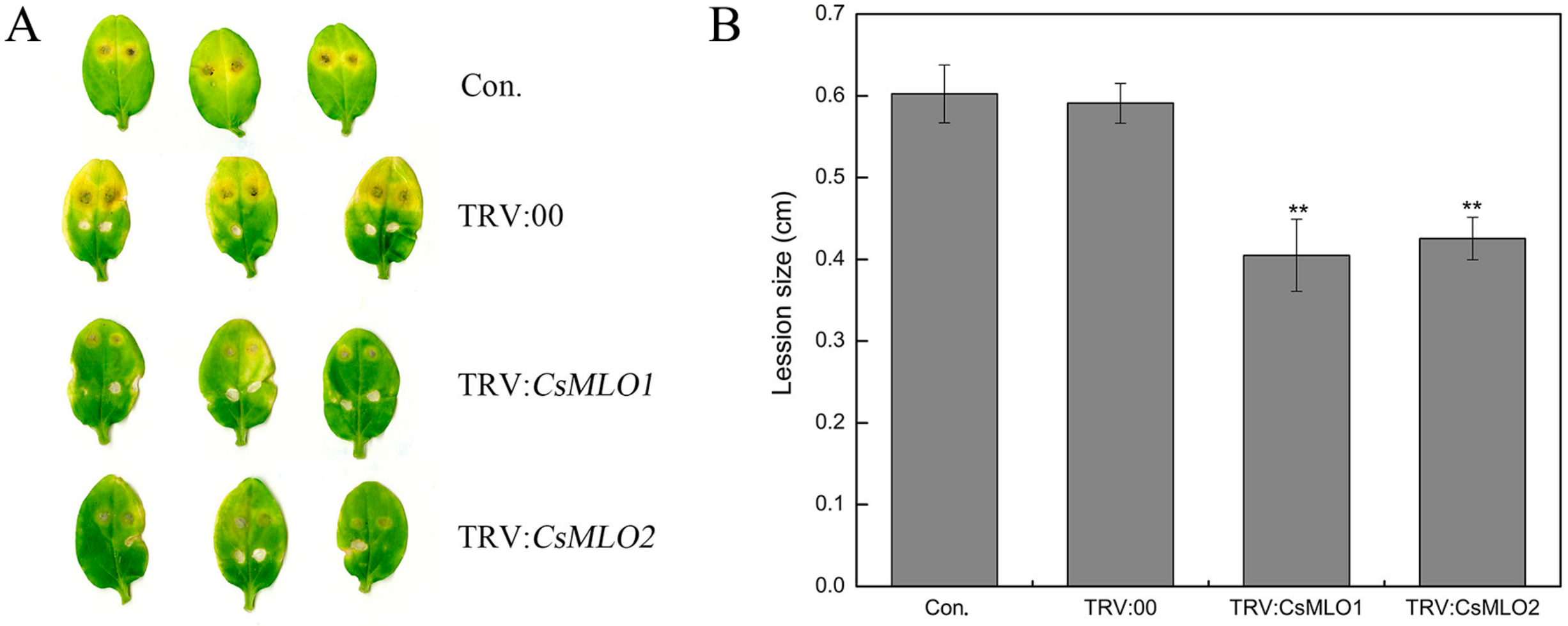
The defense resistance of CsMLO1-silenced and CsMLO2-silenced cucumbers cotyledon was evaluated following C. cassiicola inoculation for 5 d in transgenic cucumbers (Figure 4). To confirm the role of candidate genes in the defense response of C. cassiicola, we measured the lesion size caused by the pathogen infection. Based on observations and lesion-size measurement results, we found that the sizes of the lesions formed in the cucumber leaves of the non-injected cucumber cotyledons and TRV:00-injected cucumber cotyledons were equivalent. However, the lesion area of the CsMLO1/CsMLO2-silenced cucumber cotyledons was significantly smaller than that of the non-injected cucumber cotyledons and TRV:00-injected groups. The results suggested that CsMLO1-silenced or CsMLO2-silenced enhanced disease resistance to C. cassiicola in cucumber.
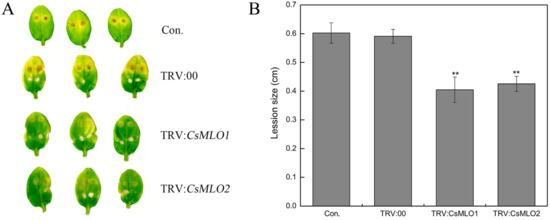
Figure 4.
Identification of disease-resistance of silenced plants after C. cassiicola inoculation. (A) The lesion areas were observed in CsMLO1-silenced and CsMLO2-silenced cucumber cotyledons after C. cassiicola inoculation; (B) Lesion size was measured in CsMLO1-silenced and CsMLO2-silenced cucumber cotyledons after C. cassiicola inoculation. Data are means ± standard deviations from three biological replicates per cultivar. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01).
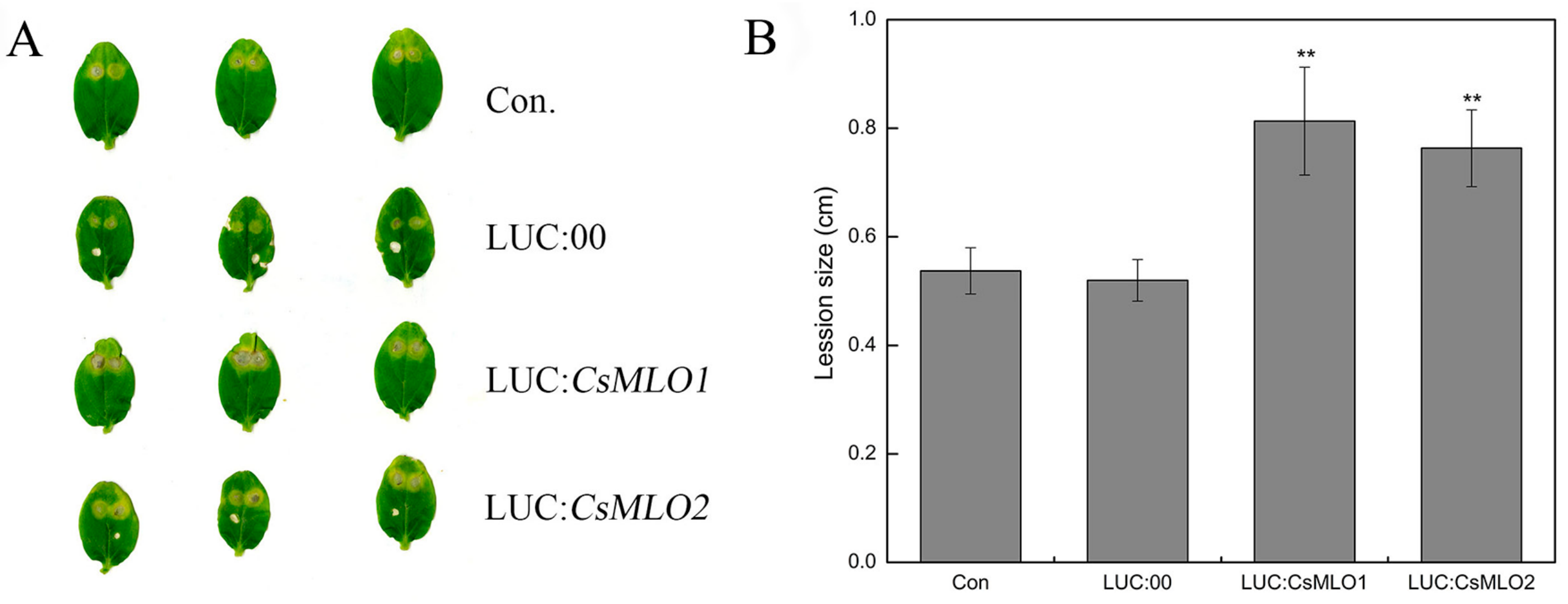
2.7. Transient Overexpression of CsMLO1 or CsMLO2 in Cucumber Cotyledons Impairs Resistance to C. cassiicola
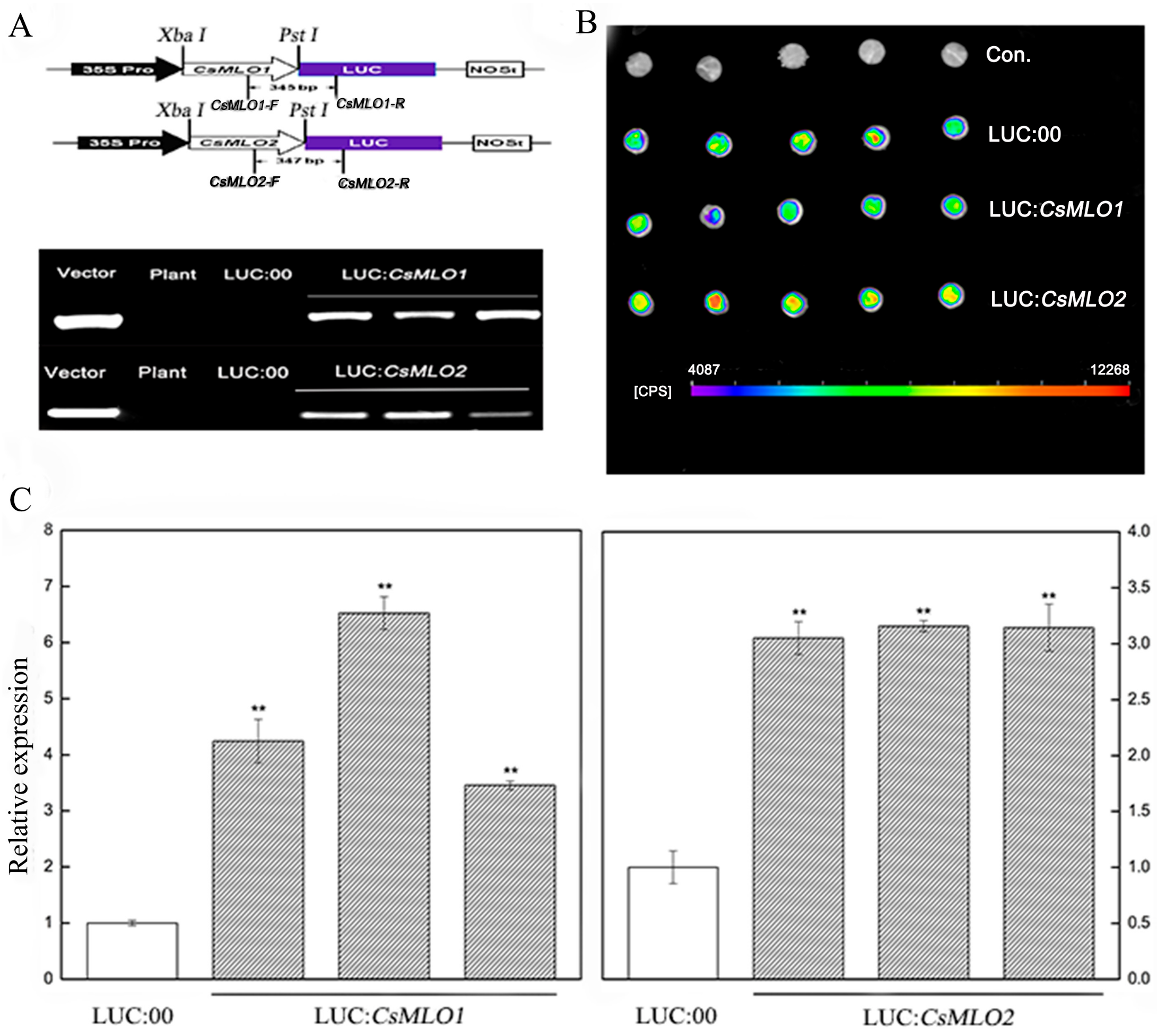
To further investigate whether the defense responses were affected by CsMLO1/CsMLO2 in cucumber cotyledons, we constructed overexpression vectors containing CsMLO1 and CsMLO2 fused with luciferase (LUC). Agrobacterium carrying LUC:CsMLO1, LUC:CsMLO2, and LUC:00 was infiltrated into the cotyledons of susceptible cucumber (Figure 5). The PCR analysis showed that the introduced genes could be detected in transient overexpression plants by chimeric primers between the CsMLO1/2 and LUC fusion genes (Figure 5A). A firefly luciferase (LUC) complementation imaging assay confirmed the success of the transient overexpression of CsMLO1/2 in vivo (Figure 5B). Strong luminescence signals were detected in the CsMLO1-LUC and CsMLO2-LUC coexpression regions but not in the non-injected cucumber cotyledons. Furthermore, the RT-qPCR assay demonstrated that the expression of the CsMLO1/CsMLO2 genes was enhanced compared with that of the LUC:00-injected cucumber cotyledons (Figure 5C). These results indicated that CsMLO1 and CsMLO2 have been successfully transiently overexpressed in cucumber cotyledons. The defense resistance levels of CsMLO1/CsMLO2-overexpressing cucumber cotyledons were assessed in cotyledons inoculated with C. cassiicola for 5 d. Increased lesion areas were observed in CsMLO1/CsMLO2-overexpressing plants compared with the non-injected and LUC:00-injected plants (Figure 6). Therefore, these results suggested that CsMLO1-overexpressing or CsMLO2-overexpressing in Xintaimici plants showed impaired resistance to C. cassiicola.
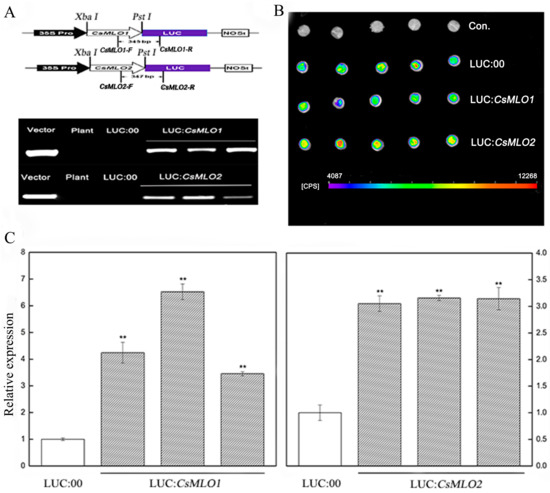
Figure 5.
CsMLO1- and CsMLO2-transient overexpressing in cucumber cotyledons. (A) Schematic of the CsMLO1-Luc and CsMLO2-Luc constructs. CsMLO1 and CsMLO2 were between 35S Pro and Luc protein, and chimeric PCR identifications of CsMLO1 and CsMLO2 genetically modified of cucumber were successful. Vector, recombinant plasmid; Plant, no-transgenic cucumber; LUC:00, empty vector; LUC:CsMLO1, CsMLO1-transient overexpressing in cucumbers; LUC:CsMLO2, CsMLO2-transient overexpressing in cucumbers. (B) Luminescence signals were detected in the CsMLO1-Luc and CsMLO2-Luc co-expression region but not in the negative controls in cucumber cotyledons; (C) Transgenic plants were identified by RT-qPCR. Data are means ± standard deviations from three independent experiments, and each column represents a sample containing three of cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01).
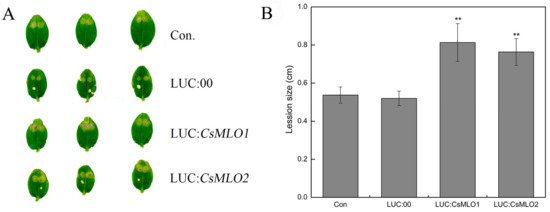
Figure 6.
Identification of disease-resistance of transient overexpressing plants after C. cassiicola inoculation. (A) The lesion areas were observated in CsMLO1- and CsMLO2-transient overexpressing cucumber cotyledons after C. cassiicola inoculation. (B) Lesion sizes were measured in CsMLO1- and CsMLO2-transient overexpressing cucumber cotyledons after C. cassiicola inoculation. Data are means ± standard deviations from three biological replicates per cultivar. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01).
2.8. Transgenic Plants Show a Defense Response after C. cassiicola Challenge
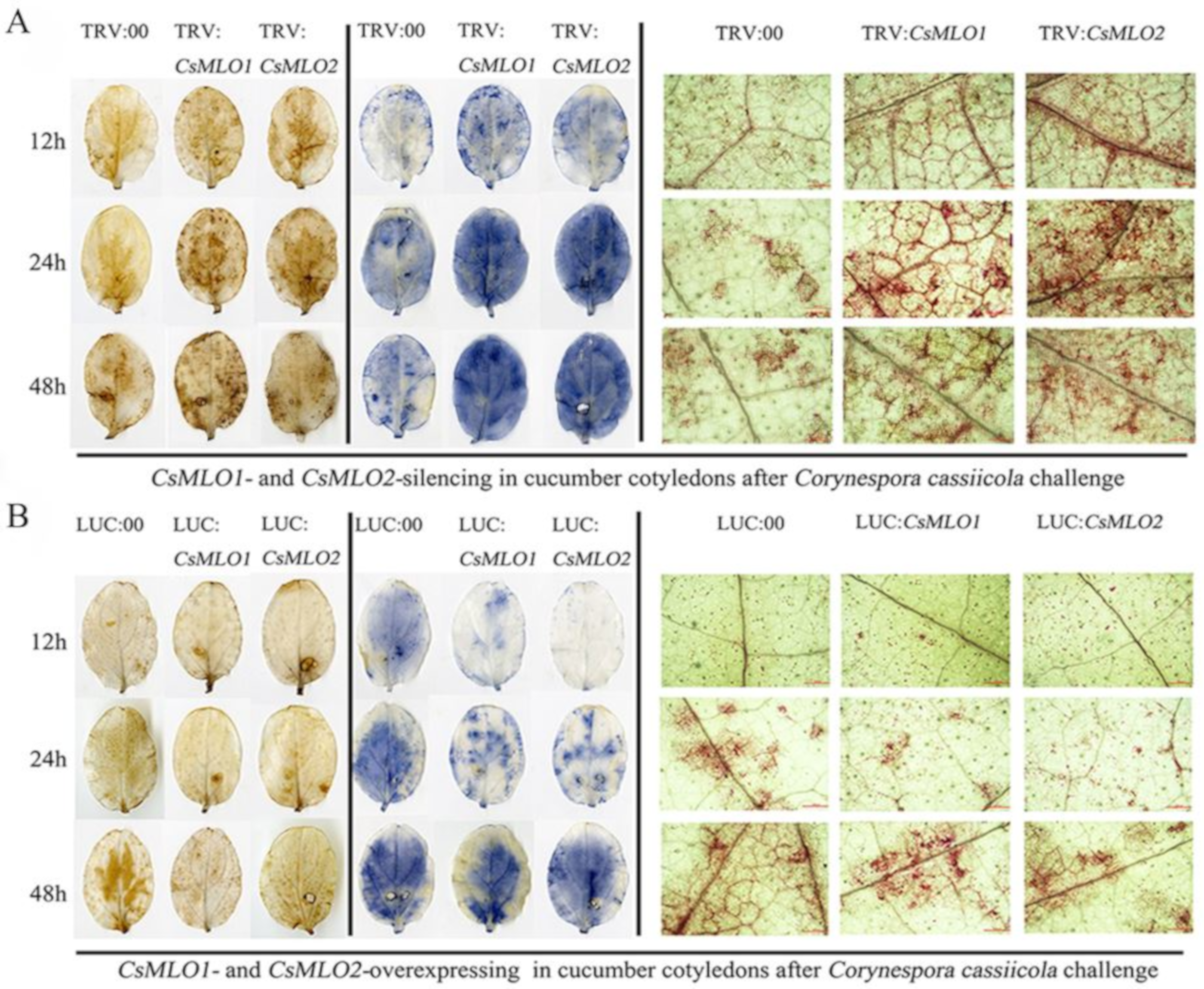
Based on the previous analysis, we speculated that the gene expression of CsMLO1 and CsMLO2 was related to ROS. Detached leaves from the transient overexpression and silenced plants subjected to C. cassiicola were used to measure the accumulation of H2O2 and O2·− using the 3,3′-diaminobenzidine (DAB) and nitrotetrazolium (NBT) staining methods, respectively (Figure 7). DAB staining showed that brown spots began to appear in CsMLO1/CsMLO2-silenced plants at 12 hpi (hours post-inoculation), which indicated that H2O2 began to accumulate compared to that in TRV:00 plants. At 24 and 48 hpi, a large area in the leaves was stained, showing that the leaves accumulated high levels of H2O2, which began to weakly decrease at 48 hpi (Figure 7A). However, at 24 hpi, brown spots began to appear in the CsMLO1/CsMLO2-overexpressing plants compared to the silenced plants (Figure 7B). At 48 hpi, deeply colored leaves and H2O2 accumulation were observed in CsMLO1/CsMLO2-overexpressing plants (Figure 7B). In the NBT staining experiment, blue spots began to appear at 12 hpi and large areas of blue began to appear in CsMLO1/CsMLO2-silenced plants at 24 and 48 hpi, whereas the O2·− accumulation began to weakly decrease at 48 hpi compared with 24 hpi (Figure 7A). However, blue spots began to appear at 24 hpi, and large areas of blue began to appear in CsMLO1/CsMLO2-overexpressing plants at 48 hpi (Figure 7B). Thus, outbreaks of ROS in silenced plants appeared earlier than those of the transient overexpression plants following C. cassiicola challenge, suggesting that the silencing of CsMLO1/CsMLO2 inhibited pathogen invasion or effectively initiated the defense responses. Moreover, histochemical staining of detached leaves at different treatment times showed lignin deposition (Figure 7). The accumulation of lignin was observed in CsMLO1/CsMLO2-silenced plants at 12, 24 and 48 h of C. cassiicola challenge (Figure 7A). Maximum lignin deposition was found in CsMLO1/CsMLO2-overexpressing cucumber cotyledons at 48 h following C. cassiicola challenge (Figure 7B). Similarly, silenced plants with C. cassiicola infection showed lignin deposition, and this change appeared earlier than that in the transient overexpression plants. We speculated that the strengthening of the cell wall in silenced plants following infection enhanced resistance and prevented pathogen invasion earlier than that in transient overexpression plants.
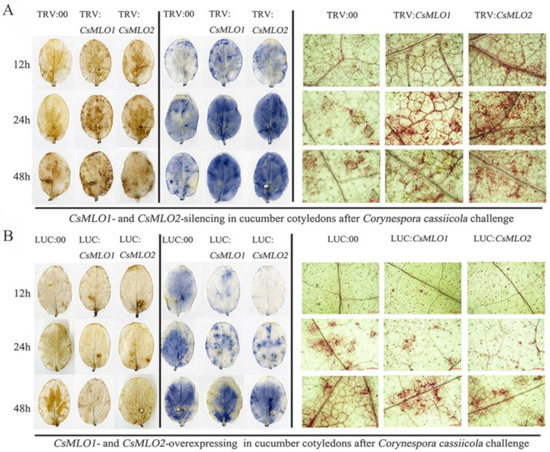
Figure 7.
Vital staining of different times were surveyed in transgenic cucumber cotyledons after treatment with C. cassiicola. (A) The accumulation of H2O2 and O2·− using the 3,3′-diaminobenzidine (DAB) and nitrotetrazolium (NBT) staining methods showed dye accumulation after treatment with C. cassiicola in silencing plants. Cell wall reinforcement showed dye accumulation after treatment with C. cassiicola by lignin staining. (B) The accumulation of H2O2 and O2·− using the DAB and NBT staining methods showed dye accumulation after treatment with C. cassiicola in transient overexpressing plants. Cell-wall reinforcement showed dye accumulation after treatment with C. cassiicola by lignin staining. Data are means three biological replicates per cultivar. Images of lignin staining were obtained using a Nikon optical microscope. Scale bars = 500 μm.
2.9. CsMLO1 or CsMLO2 Transgenic Plants Modulate the Expression Levels of Defense-Related Genes
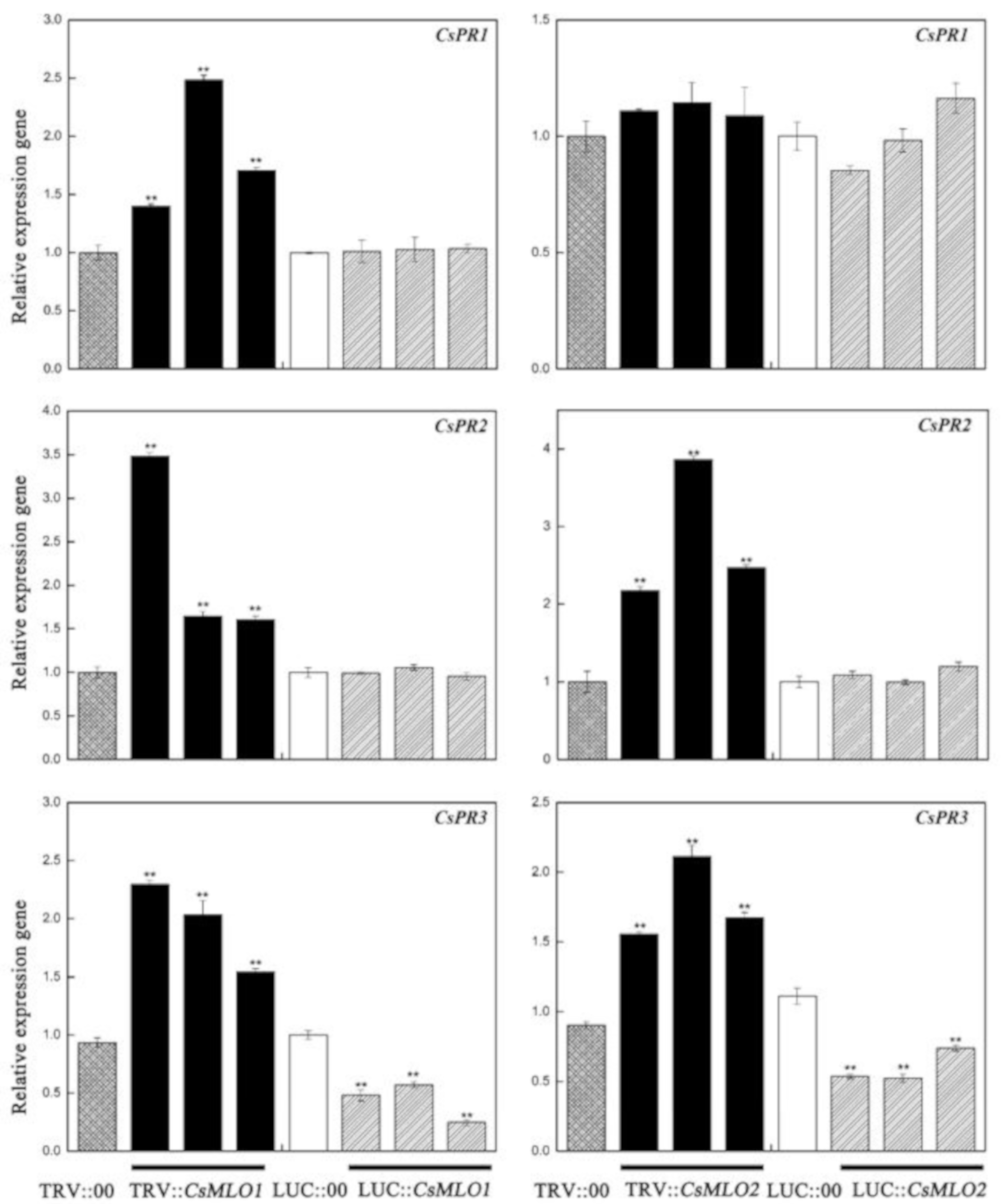
To explore whether CsMLO1/CsMLO2-modulated expression influenced disease resistance against C. cassiicola, we analyzed cucumber pathogenesis-related proteins, including PR1-1a (AB698861), PR2 (XM_011661051), and PR3 (HM015248), in transgenic cucumbers (Figure 8). In our experiment, the transcript level of PR1-1a was only upregulated in CsMLO1-silenced cucumber cotyledons. The expression levels of the PR2 and PR3 genes were increased in CsMLO1-silenced and CsMLO2-silenced cucumber cotyledons compared with those of the TRV:00 plants. However, the transcript levels of PR2 and PR3 showed differing degrees of variance and declined in the transient overexpression plants. Based on the above results, we speculate that the CsRP might be involved in the defense pathway by CsMLO1/CsMLO2-induced defense pathways, whereas the CsMLO1-induced defense pathway might be related to the CsRP1-1a gene.
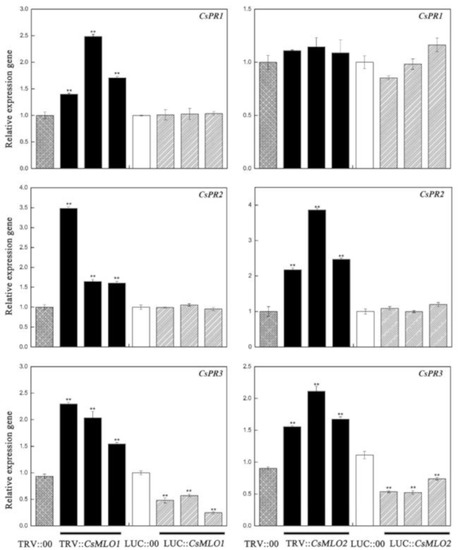
Figure 8.
Effect of transgenic expressions of CsMLO1 and CsMLO2 in cucumber cotyledons on immunity induction. The different expression regulations of pathogenesis-related proteins (CsPR1-1a, CsPR2 and CsPR3) are shown in transgenic plants. Data are means ± standard deviations from three independent experiments, and each column represents a sample containing three of cucumber cotyledons from different plants. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01).
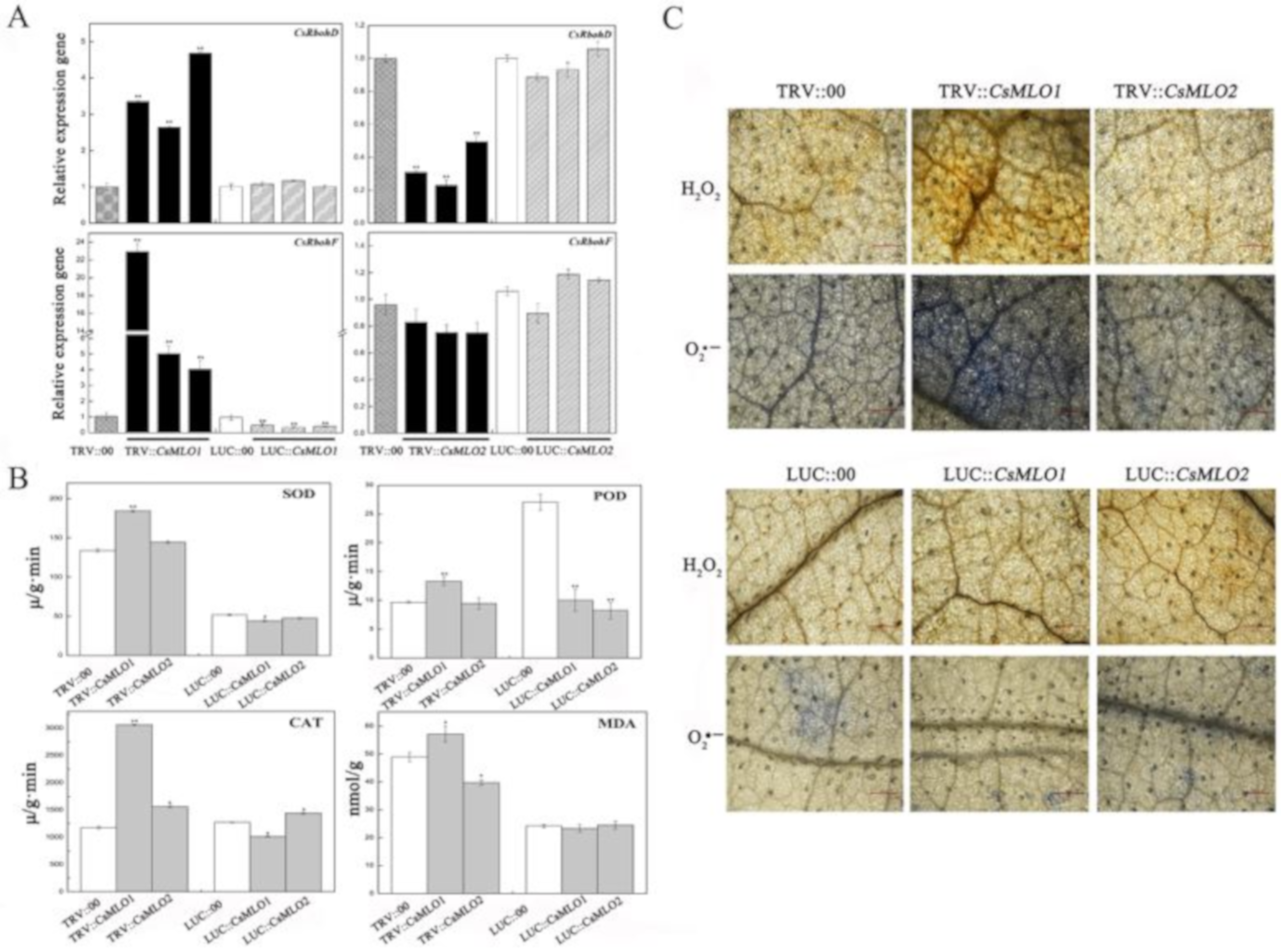
2.10. CsMLO1 Transient Transgenic Plants Modulate the Expression Levels of Reactive Oxygen Species (ROS)-Related Genes and Abscisic Acid (ABA) Signaling-Related Genes
The above study demonstrated the involvement of CsMLO1 and CsMLO2 responses to exogenous H2O2 in cucumber. Next, we investigated whether the ROS formation-related genes were affected in CsMLO1 and CsMLO2 transient transgenic plants (Figure 9). The transcript levels of the CsRbohD (Cucsa.340760) and CsRbohF (Cucsa.107010) genes were significantly increased in CsMLO1-silenced plants and decreased in the CsMLO1-overexpressing plants compared to those of the controls. However, CsRbohD and CsRbohF expression declined in CsMLO2-silenced plants (Figure 9A). Thus, CsMLO1 silencing promoted ROS formation. To further verify these results, we examined antioxidant enzymes in CsMLO1 and CsMLO2 transient transgenic plants (Figure 9B). The activities of total superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) were higher in CsMLO1-silenced plants than in control plants but were lower in CsMLO1-overexpressing plants than control plants. Subsequently, excess ROS degraded polyunsaturated lipids to form malondialdehyde (MDA); the MDA level was significantly increased in CsMLO1-silenced plants, and opposite results were obtained in CsMLO1-overexpressing plants. However, the activities of these enzymes were decreased or unchanged in CsMLO2-transgenic plants and did not show a regular pattern. In addition, DAB and NBT staining was used to detect H2O2 and O2·− in CsMLO1 and CsMLO2 transgenic plants (Figure 9C). After CsMLO1 silencing, the H2O2 and O2·− levels were significantly increased, resulting in brown and blue spots that were more intense than those in the TRV:00 leaves, whereas the leaf color was not obviously changed in the CsMLO1-overexpressing plants compared with that in the LUC:00 plants. The DAB and NBT staining in all examined CsMLO2 transient transgenic cucumber cotyledon was similar to that of the TRV:00 and LUC:00 leaves. Based on these results, the clear signal pathways between CsMLO1 and ROS were further confirmed. The CsMLO1-silenced plants triggered ROS signaling in cucumber cotyledons as a defense response to C. cassiicola.
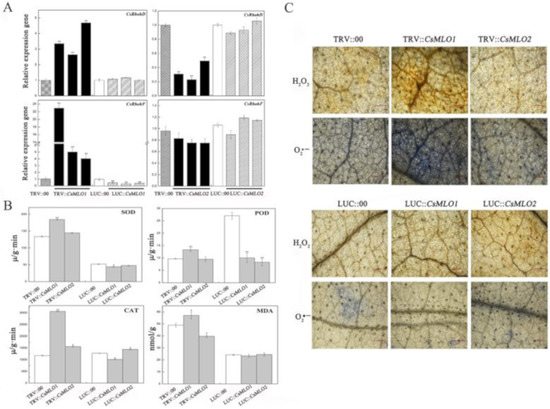
Figure 9.
CsMLO1 and CsMLO2 transgenic plants modulate the ROS signaling. (A) The transcript level of CsRbohD and CsRbohF were detected by RT-qPCR in CsMLO1/CsMLO2-overexpressing and CsMLO1/CsMLO2-silencing cucumber cotyledons. Data are means ± standard deviations from three independent experiments, and each column represents a sample containing six of cucumber cotyledons. Expression analysis of candidate genes using the 2−ΔΔCt method. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01). (B) Antioxidant enzymes were examined in CsMLO1 and CsMLO2 transgenic plants. Data are means ± standard deviations three biological replicates per cultivar. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01); (C) ROS-related (H2O2 and O2·−) staining were shown in CsMLO1/CsMLO2-overexpressing and CsMLO1/CsMLO2-silencing cucumber cotyledons. Data are means three biological replicates of per variety. Images of staining were obtained using a Nikon optical microscope. Scale bars = 500 μm.
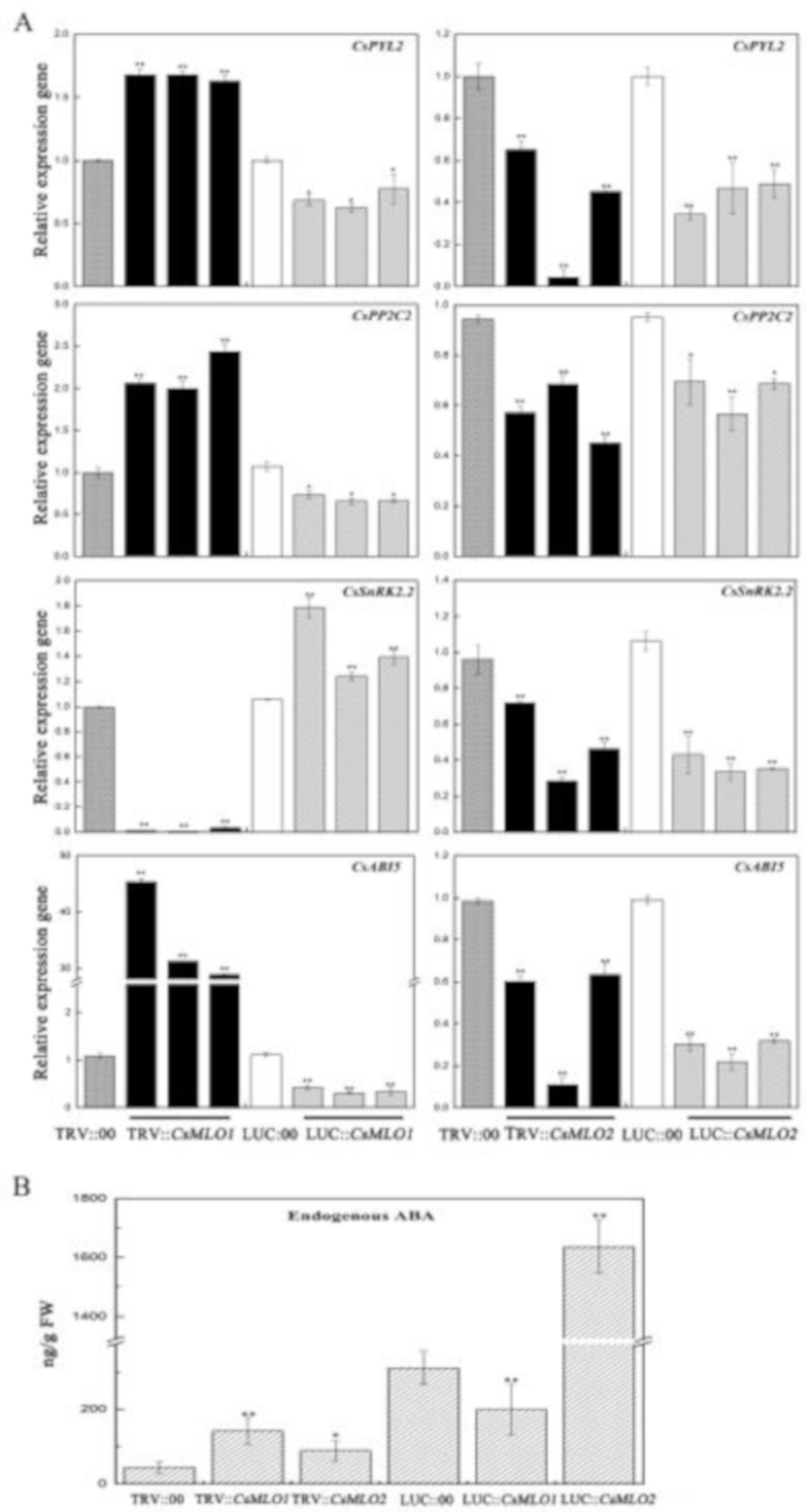
As mentioned above, the transcript levels of CsMLO1 and CsMLO2 responded to exogenous ABA in cucumber. We next assayed the role of CsMLO1 and CsMLO2 in the regulation of endogenous ABA signaling-related genes in transient transgenic cucumber cotyledon (Figure 10). Compared with the control levels, the transcript levels of CsPYL2 (JF789829) and CsPPC2 (JN566067) were significantly increased in CsMLO1-silenced plants (Figure 10A) and decreased in the CsMLO1-overexpressing plants; however, the transcript levels of CsPYL2 and CsPPC2 were reduced in the CsMLO2-silenced and CsMLO2-overexpressing plants. SnRK2.2 (JN566071) expression was almost eliminated in the CsMLO1-silenced cucumbers, and a decreasing pattern also appeared in the CsMLO2-silenced/overexpressing plants. The transcript level of SnRK2.2 was increased in the CsMLO1-overexpressing cucumbers. In contrast, CsABI5 (XM_004149176.2) gene transcription was upregulated at least 30-fold in the CsMLO1-silenced cucumbers compared with the TRV:00-injected cucumbers and slightly decreased in the CsMLO1-overexpressing cucumbers. In addition, the transcript levels of CsABI5 were markedly lower in the CsMLO2-silenced/overexpressing plants than in the TRV:00/LUC:00-injected plants. Our data suggested that CsMLO1 negatively modulated the expression of CsPYL2, CsPP2C2 and CsABI5 and positively modulated the expression of CsSnRK2.2. Based on these findings, we next analyzed the accumulation of endogenous ABA via ELISAs of the cotyledons of transient transgenic cucumbers (Figure 10B). To experimentally verify the above results, we assessed the accumulation of ABA, which was enhanced in the CsMLO1-silenced plants compared with TRV:00 plants, whereas the accumulation of ABA was reduced in the CsMLO1-overexpressing plants compared with LUC:00 plants. The accumulation of endogenous ABA was increased in the CsMLO2-silenced/overexpressing plants. Together, these results suggested that CsMLO1 was a negative regulator of ABA-induced gene expression in the cucumber cotyledons, thus modulating the defense response to C. cassiicola. Although CsMLO1 and CsMLO2 belong to the same family and were highly similar, they may have different mechanisms of action against pathogens.
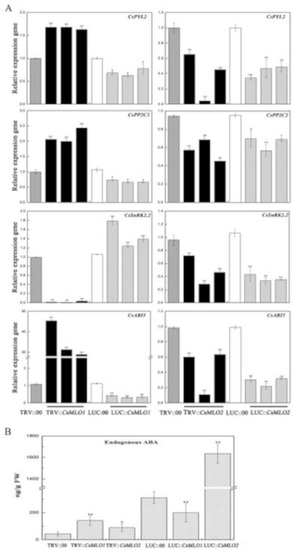
Figure 10.
CsMLO1 and CsMLO2 transgenic plants modulate the ABA signaling. (A) ABA-related genes (CsPYL2, CsPP2C2, CsSnRK2.2 and CsABI5), were observed in transgenic plants by RT-qPCR. Expression analysis of candidate genes using the 2−ΔΔCt method. (B) The accumulation of endogenous ABA was tested in the CsMLO1/CsMO2-silencing and CsMLO1/CsMO2-overexpressing in cucumber cotyledons. Values in (A) and (B) are means ± standard deviations from three independent experiments, and each column represents a sample containing six of cucumber cotyledons. The asterisks indicated a significant difference (Student’s t-test, *P < 0.05 or **P < 0.01).
3. Discussion
In this study, CDS regions of two MLO genes, namely, CsMLO1 and CsMLO2, were cloned and identified. A sequence analysis showed that the CsMLO1 and CsMLO2 proteins had seven typical transmembrane domains and a CaMBD, and these domain modules were similar to those of CaMLO1, AtMLO2, AtMLO6, BrMLO1 and HvMLO1 [14,44,45]. The cucumber CaMBD1 domain of CsMLO1 interacts with CsCaM3 via yeast two-hybrid, firefly luciferase (LUC) complementation and bimolecular fluorescence complementation (BiFC) experiments [46]. Plasma membrane proteins may participate in defense mechanisms against biological stress [47]. Studies of barley mlo mutant plants have indicated that MLO proteins play a negative regulatory role in the defense response [14,48]. Three closely related MLO co-orthologs (AtMLO2, AtMLO6, and AtMLO12) in clade V were mutated to achieve complete resistance to Golovinomyces orontii [16,49]. In our study, a phylogenetic analysis showed that a similar number of MLO family genes were predicted in cucumber and CsMLO1 and CsMLO2 were classified into the same clade as AtMLO2, AtMLO6, AtMLO12, BrMLO1 and CaMLO1. The results indicated that loss of function of CsMLO1 and CsMLO2 could play a role in cucumber defense. Pepper CaMLO2 is localized in the plasma membrane and involved in the susceptibility cell-death response and bacterial and oomycete proliferation [43]. A subcellular localization analysis showed that the CsMLO1::GFP and CsMLO2::GFP fusion proteins appeared solely in the plasma membrane as fusion proteins in Nicotiana benthamiana leaf cells. These results were consistent with the localization of CsMLO1 and CsMLO2 in cucumber protoplasts [46]. Therefore, it was further demonstrated that these results were consistent with the subcellular localization results of most MLO disease-resistance genes, and CsMLO1 and CsMLO2 showed potential functions in the plasma membrane.
Traditionally, MLO function has been associated with susceptibility/resistance to powdery mildew (PM) disease [13]. A deletion variant (∆174) of CsaMLO8 has lost its function as a susceptibility gene, thus leading to PM resistance in complementation of the tomato mlo-mutant [50]. In recent years, studies have also found that the MLO gene is involved in regulating other pathogens [17,18,19,20,21,22,23]. In cucumber–C. cassiicola interactions, the role of the MLO gene is unclear. Here, we found that the lesion area was significantly higher in susceptible cucumbers than in resistant cucumbers. CsMLO1 expression was upregulated in resistant and susceptible cultivars challenged with C. cassiicola. Compared with the resistant cultivar at 48 h, the susceptible cultivar showed highly upregulated CsMLO1 expression. CsMLO2 expression was initially downregulated in the resistant and susceptible cultivars challenged with C. cassiicola and then upregulated at 48 h. Similarly, CsMLO2 expression was highly upregulated in the susceptible cultivar. These results suggested that the maximum accumulation of CsMLO1 and CsMLO2 in the susceptible cultivar appeared earlier than that in the resistant cultivar in the defense response to C. cassiicola. Currently, due to the limitation of cucumber transgenic technology, research on the disease-resistance mechanism of the MLO gene has been hampered in cucumber. However, an experimental method for the transient agroinfiltration of cucumber cotyledons has been established [51]. In this study, we first successfully obtained transgenic cucumbers that showed transient silencing and overexpression of CsMLO1/CsMLO2. Next, a functional analysis revealed that CsMLO1/CsMLO2-silenced plants showed strongly enhanced resistance to C. cassiicola, although CsMLO1/CsMLO2-overexpressing plants showed evidently impaired resistance to C. cassiicola in the transient transgenic cucumbers. ROS accumulation and lignin deposition also occurred earlier in CsMLO1/CsMLO2-silenced cucumbers than CsMLO1/CsMLO2-overexpressing cucumbers after C. cassiicola infection. Wang has shown that secondary metabolism and ROS accumulation play important roles in disease resistance during cucumber-C. cassiicola interactions [25]. Thus, we initially concluded that CsMLO1 and CsMLO2 might be negative regulatory modulators involved in the cucumber defense response to C. cassiicola.
In Arabidopsis, clade V MLOs act as negative regulators of ROS signaling, which suggests that ROS signaling is a general feature of MLO proteins [30,52,53]. Normally, plants trigger ROS signaling in response to both abiotic and biotic stresses. In cucumber, CsMLO1 and CsMLO2 were classified into the same clade as clade V AtMLOs. Thus, cucumber CsMLO1 and CsMLO2 are likely associated with ROS signaling in response to C. cassiicola. In addition, high concentrations of ROS chemicals trigger plant HR against the invasion of pathogens [54,55]. AtrbohD and AtrbohF are associated with the production of ROS in response to ABA and pathogen infection, such as Pseudomonas syringae and Hyaloperonospora arabidopsis [31,32,56]. In our study, we found that transcript levels of CsMLO1 were suppressed by treatment with H2O2, whereas transcript levels of CsRbohD and CsRbohF were increased in CsMLO1-silenced cucumbers. Meanwhile, increased antioxidant enzymes and H2O2 and O2−· accumulation were observed in CsMLO1-silenced cucumbers. Similar results were observed in an earlier study of the response to ROS, which was identified using publicly available gene expression data of the mlo2-6 mlo6-2 mlo12-1 mutant [17,30]. This pattern suggested that ROS signaling might be preactivated in the CsMLO1-silencing cucumber to improve defense resistance to C. cassiicola.
The calcium signal and the reactive oxygen species signal are inseparable in plants. Ca2+ influx can induces the production of reactive oxygen species, which can activate Ca2+ influx. Cytosolic Ca2+ transients modulate RbohD- and RbohF-mediated ROS accumulation as well as calmodulin (CaM)-mediated defense signaling [57,58,59,60,61]. ROS can act as secondary signals in the activation of a series of downstream pathogenesis-related proteins [62]. Cucumber CsCaM3 is a positive modulator that enhances the defense response of C. cassiicola infection, but CsMLO1 is a negative modulator to enhance the defense response of cucumbers and stably interacts with CsCaM3 and transfers CsCaM3 in the cytoplasm to the plasma membrane, thereby blocking the accumulation of CsCaM3 [41]. Cucumber CsMLO1 silencing significantly enhances the expression of reactive oxygen species (ROS)-related genes (CsPO1, CsRbohD, and CsRbohF), defense marker genes (CsPR1 and CsPR3), CsCaM3 and callose deposition-related gene (CsGSL) under C. cassiicola infection [41]. Additionally, the coexpression of CsCaM3 and CsMLO1 significantly inhibited hypersensitive cell death after C. cassiicola infection, suggesting that CsMLO1 negatively regulates CsCaM3 expression, which results in the inhibition of defense-related gene activation. In our study, the transcript level of PR1-1a was only increased in CsMLO1-silenced cucumbers. The transcript levels of PR2 and PR3 were significantly increased in the CsMLO1/CsMLO2-silenced cucumbers compared with the TRV:00-injected cucumbers, whereas PR3 expression was reduced in the CsMLO1/CsMLO2-overexpressing cucumbers. These results are consistent with the results of Xue, who reported that the enhanced expression of defense-related genes improved disease resistance against B. cinerea in Arabidopsis [63]. The CsMLO1 gene and CsMLO2 gene belong to the same family, and they are involved in the same defense pathways against pathogens, such as active oxygen bursts and lignin deposition; however, different regulatory mechanisms are observed, such as activation of different pathogenesis-related proteins. Different expression patterns of PR proteins also indicated the possible genetic redundancy between CsMLO1 and CsMLO2 paralogs in the regulation of PR genes. In summary, CsMLO1 silencing activates calcium signaling and reactive oxygen species signaling pathways, and crosstalk between Ca2+ signaling and ROS signals may jointly activate downstream pathogenesis-related gene expression and antioxidant enzyme activity. Negative regulatory modes of CsMLO2 in cucumber might be related to PR protein defense against C. cassiicola.
In plants, complex signaling transduction pathways occur in plant–pathogen interactions [64]. The modulation of ABA signaling plays a crucial role in plant stress responses. An example of virus-induced silencing in pepper plants suggested that CaMLO2 acts as a negative regulator of ABA signaling in drought stress responses [65]. In this study, we detected the expression of CsPYL2, CsPP2C2 and CsSnRK2.2, which initiate ABA signal transduction, and found that CsMLO1 negatively modulated the expression of PYL2 and CsPP2C2 and positively modulated the expression of SnRK2.2. Moreover, endogenous ABA also showed the same trend as the gene expression in CsMLO1-silenced/overexpressing cucumbers. Thus, we speculated that CsMLO1 might be involved in and regulate the ABA signaling pathway. ABA is an important signaling molecule in plant–pathogen interactions [66,67,68]. In this study, a functional analysis showed that CsMLO1 silencing significantly increased the expression of CsABI5, although the transcript level of CsABI5 was inhibited in CsMLO1-overexpressing cucumbers. Changes in the expression levels of CsABI5 directly demonstrated the involvement of CsMLO1 in ABA signaling in defense responses. Furthermore, these results were relatively similar to the augmented CsMLO1 response to C. cassiicola, indicating that CsMLO1-silencing signals mediated and enhanced the response against the pathogen. Previous evidence showed that ABA levels were correlated with resistance to pathogen stress and that ABI5 was highly induced in a resistant line compared with a susceptible line after S. fuliginea infestation [69]. These results are also consistent with the response of CsMLO1 to C. cassiicola, which was associated with the ABA signaling pathway. The research find that ABA stimulate the influx of calcium ions in the leaves of maize seedlings, which in turn increased the activity of plasma membrane NADPH oxidase and the production of O2−·, resulting in an increase in ROS content [70]. ABA and H2O2 can activate Ca2+-CaM targets and upregulate antioxidant enzyme activity [71,72,73]. Together, these findings suggest that the functional role of CsMLO1 in the defense response to C. cassiicola was also correlated with ABA signaling. The CsMLO1 silencing increased the content of endogenous ABA and upregulated the expression of related genes in cucumber leaves, and activated the ROS signal and Ca2+-CaM signaling pathway. However, the above assays found that the transcription level of CsMLO2 was affected by exogenous ABA while the expression of ABA-related genes showed a downward trend compared with that of the control in CsMLO2-silenced/overexpressing cucumbers. These findings further suggest that although CsMLO2 also regulated ABA signaling, whether this signaling is involved in defense against C. cassiicola remains unclear.
In conclusion, the cucumber CsMLO1 and CsMLO2 genes were identified as negative regulators of the defense response to C. cassiicola. Combined with the previous studies and the results of this study, it was found that CsMLO1 as a negative regulator mainly adopted the following resistance pathways. CsMLO1-silenced mainly activated the expression of Ca2+-, ROS-, ABA- signaling-related genes, thereby improving the resistance of cucumber to C. cassiicola by regulating crosstalk between the three signaling; The interaction of CsMLO1 and Ca2+-CaM inhibited HR response mainly including ROS burst, CaM and pathogenesis-related gene expression. Furthermore, cucumber CsMLO2 regulated the expression of some pathogenesis-related proteins, which influenced the defense response to C. cassiicola. Taken together, these studies will provide novel insights into the important roles of CsMLO1 and CsMLO2 in the defense responses to C. cassiicola.
4. Materials and Methods
4.1. Plant Materials and Treatments
The cucumber cultivars Jinyou 38, B21-a-2-1-2, B21-a-2-2-2, F10, 995, Jinyan 4 and Xintaimici were used in this study. Jinyou 38 is an important planting variety in China that is highly resistant to C. cassiicola. B21-a-2-2-2 (highly susceptible to S. fuliginea) and B21-a-2-1-2 (highly resistant to S. fuliginea) are two sister cucumber lines. F10 (highly resistant to Fusarium axysporum) and 995 (highly susceptible to Fusarium oxysporum) were obtained from the Liaoning Academy of Agricultural Sciences. JingYan4 is resistant to downy mildew and C. cassiicola in China. Xintaimici is susceptible to C. cassiicola and was selected for gene transformation. The cucumbers were sown in peat soil-vermiculite (1:2, v/v) under 25 °C, a 16 h light/8 h darkness cycle, and 60% relative humidity.
4.2. Pathogen Growth and Inoculation
C. cassiicola was obtained from the species conservation center of the Chinese Academy of Agricultural Sciences, and it was streaked on PDA solid medium and then stored in an incubator at 25 °C. The spore suspensions were harvested in sterile water and adjusted to a final concentration of 2 × 105 sporangia/mL. The spore suspensions were sprayed onto cotyledons of transgenic cucumbers. Inoculated cucumbers were covered with plastic to maintain moisture and grown at 25 °C in a growth chamber. Then, 3-week-old cucumber seedlings of cucumber strains were inoculated at the second leaf stage with C. cassiicola, and control plants were inoculated with distilled water. The disease progression of corynespora leaf spot was estimated on the basis of the severity of leaf scabs as follows: Grade 0: no scabs observed; Grade 1: Less than 1/10 of leaves infected; Grade 3: 1/10–1/4 of leaves infected; Grade 5: 1/4–1/2 of leaves infected; Grade 7: 1/2–3/4 of leaf infected; and Grade 9: More than 3/4 of leaves infected. Disease index = 100 × ∑ (no. of diseased leaves of each grade × disease grade)/(total no. leaves × 9).
4.3. Analysis of Sequence and Phylogenetic Tree
The nucleotide and deduced amino acid sequences of CsMLO1 and CsMLO2 cDNA that encode MLO homolog proteins were analyzed, and a phylogenetic tree was generated using 19 resistant proteins of known function.
4.4. Subcellular Localization
The RNA of the leaves was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China), and synthesized into cDNA using the FastQuant RT Kit (Tiangen, Beijing, China). The CsMLO1 and CsMLO2 cDNA regions were amplified using PrimeSTAR GXL DNA Polymerase (Takara, Dalian, China). For the PCR analysis, the following cycling parameters were used: 98 °C (10 s), 60 °C (2 min), 68 °C (30 s), 32 cycles in total. The PCR products were detected by agarose gel electrophoresis. The PCR products were introduced into pRI101-GFP vector to form two constructs with the p35S::CsMLO1-GFP fusion gene and p35S::CsMLO2-GFP fusion gene. Recombinant plasmids were transformed into Agrobacterium tumefaciens strain EHA 105 and then centrifuged after overnight culture. The precipitate was cultured in induction medium (10 mmol·L−1 ethanesulfonic acid, pH 5.7, 10 mmol·L−1 MgCl2 and 200 mmol·L−1 acetosyringone), harvested and diluted to OD600 = 0.8, and then injected into Nicotiana benthamiana leaves. Two days after infiltration, the epidermal cells and protoplasts were observed using a confocal laser-scanning microscope (Leica TCS SP8, Solms, Germany). The BP505-530 filter sets (excitation 488 nm, emission 505 to 530 nm) were used to detect GFP.
Protoplasts were extracted from transformed tobacco leaves. The transformed tobacco leaves were cut into 0.5–1 mm thin strips, quickly transferred to the enzymatic hydrolysate (20 mmol·L−1 2-(4-Morpholino) ethanesulfonic acid (MES), 1.5% Cellulase R-10, 0.4% Macerozyme R-10, 20 mmol·L−1 KCL, 0.4 mol·L−1 mannitol, 10 mmol·L−1 CaCl2, and 0.1% bovine serum albumin (BSA), pH 5.7), and after 30 min of vacuum infiltration, shaken at 50 rpm for 6 h. The enzymatic hydrolysate was diluted with an equal amount of W5 (0.2 mol·L−1 MES, 1.54 mol·L−1 NaCl, 1 mol·L−1 CaCl2, and 0.2 mol·L−1 KCl, pH 5.7) and then filtered through a 100 μm nylon membrane. The collected filtrate was centrifuged at 100 × g for 2 min. The supernatant was slowly removed, and the remaining green liquid contained protoplasts. An equal amount of precooled MGG (0.2 mol·L−1 MES, 0.4 mol·L−1 Mannitol, 1.5 mol·L−1 MgCl2) was placed in the protoplasts and placed on ice for observation using a confocal laser scanning microscope.
4.5. Application of Plant Abiotic and Biotic Stress
The experimental materials included one-month-old cucumber seedlings (Jinyou 38 and Xintaimici). To simulate abiotic stress, the leaves of Jinyou 38 were sprayed with 10 μmol·L−1 H2O2, 100 μmol·L−1 ABA, 1 mmol·L−1 salicylic acid (SA), 100 μmol·L−1 jasmonate (MeJA) and 10 mmol·L−1 CaCl2. The control seedlings were sprayed with distilled water (H2O). Biotic stress, the leaves were sprayed with C. cassiicola inoculation. The control seedlings were sprayed with distilled water. Abiotic stress-treated cucumber leaves were collected at 12, 24, 48 h and the inoculated leaves were harvested at seven time points (i.e., 0, 6, 12, 24, 48, 72 and 144 h post-inoculation). The harvested leaves frozen in liquid nitrogen and then stored at –80 °C.
4.6. Quantification of Genes Expression using Reverse-Transcription Quantitative PCR (RT-qPCR)
Total RNA was extracted from cucumber euphylla and cucumber cotyledons by RNAprep Pure Plant Kit (Tisngen biotech, Beijing, China). The primers for candidate genes were designed using QuantPrime-a flexible primer design tool for high-throughput qPCR by http://quantprime.mpimp-golm.mpg.de/. RT-qPCR was performed on a SYBR Green I 96-I system (Roche fluorescence quantitative PCR instrument, Basel). Actin gene of cucumber was used as the internal reference [74]. The relative expressions of the target genes were calculated using the 2−ΔΔCT method [75]. All the primers were listed in Table S2.
4.7. Histochemical Analysis
For 3, 3’-diaminobenzidine (DAB; Sigma, St. Louis, MO, USA) and nitrotetrazolium blue chloride (NBT; Ameresco, OH, USA) to visualize H2O2 and O2·− [76], transient expressions development of CsMLO1 and CsMLO2 in cucumber cotyledons were assessed after C. cassiicola inoculation at the three time points (i.e., 12, 24, 48 h post-inoculation) by DAB and NBT staining. The infected cucumber cotyledons were soaked with in 1 mg/mL DAB for 8 h and infiltrated with 0.1% NBT for 5 h, boiled for 20 min in 3:1:1 ethanol/lactic acid/glycerol and then transferred to 95% ethanol at 4 °C stored. Three independent replicates were performed for each assay.
Lignin staining was performed to visualize the degree of cell wall thickening after pathogenic infection [77]. Transient expression cucumber cotyledons after C. cassiicola inoculation were immersed for 24 h in stationary liquid at the indicated time points. Vacuum-treated in saturated aqueous solution of chloral hydrate for 10 min at room temperature for 2 to 3 days. The leaves were stained in 1% (w/v) phloroglucinol in 92% ethanol for 10 min at room temperature. Subsequently, the tissues were mounted with HCl, and red staining was immediately monitored using Inverted microscopy (Nikon Ts2, Tokyo, Japan).
4.8. Enzyme Extraction and Antioxidant Enzyme Assays
Transgenic cucumber leaves were crushed into homogenate and were extracted in pre-chilled sodium phosphate buffer (pH 7.8) and 1% (w/v) polyvinylpolypyrrolidone (PVPP). The homogenate was centrifuged at 12,000× g at 4 °C for 20 min. Activities enzyme of antioxidant enzymes superoxide dismutase (SOD), peroxidase (POD), catalase (CAT) and malondialdehyde (MDA) were immediately determined in the supernatant [78,79].
4.9. Virus-Induced Gene Silencing (VIGS)
pTRV (tobacco rattle virus)-based VIGS was performed to knock down the CsMLO1 gene and CsMLO2 gene in cucumber cotyledons. A 402 bp fragment within the 3′ region of the CsMLO1 cDNA (nucleotides 1348–1749) and a 387 bp fragment within the 3′ region of CsMLO2 cDNA (nucleotides 1347–1725) were cloned into the pTRV2 vector (TRV:CsMLO1 and TRV:CsMLO2, respectively). The gene-specific primers are listed in Table S3.
A 10 mL culture of each A. tumefaciens strain to be used was grown overnight at 28 °C in YEP medium supplemented with 100 mg·L−1 of rifampicin and 50 mg·L−1 of kanamycin. Then, 200 μL of each overnight culture was inoculated into 20 mL portions of YEP medium with antibiotics as above and cultivated at 28 °C until the culture reached selected optical densities of OD600 = 0.8–1.0.
The induced Agrobacterium tumefaciens EHA105 strains carrying different pTRV2-derived vectors (pTRV2, pTRV2-CsMLO1 and pTRV2-CsMLO2) were mixed with the pTRV1 A. tumefaciens strain EHA105. The ratio was 1:1. The samples were supplemented with 10 mmol·L−1 MES, 10 mmol·L−1 MgCl2, and 200 μmol·L−1 acetosyringone (AS) and then coinfiltrated into fully expanded cotyledons of cucumber plants (OD600 = 0.4 for each construct). Plants were placed in a growth room at 22 °C with a 16 h light and 8 h dark photoperiod for growth [46].
4.10. CsMLO-Luciferase (LUC) Fusion Overexpression Vector
The full-length CsMLO1 and CsMLO2 cDNA sequences were constructed using the pCAMBIA 3301 vector with luciferase (LUC), and they were under the control of the CaMV 35S promoter in cotyledons of cucumbers. The gene-specific primers are listed in Table S3. The correct constructs were introduced into the A. tumefaciens strain EHA 105. Agrobacterium-meditated transformation with the LUC:00, LUC:CsMLO1 gene and LUC:CsMLO2 gene was carried out. The experimental method was the same as the above.
4.11. Statistical Analysis
Primer design and sequence alignment were conducted using Primer 5 software. Data are the mean ± standard deviation from three biological replicates per cultivar. Standard errors of deviation were assessed by Excel. Statistical significance was analyzed by Student’s t-test (P < 0.05 or P < 0.01) using SPSS software (SPSS 22.0, Nlinedown, Guangdong, China).
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/19/4793/s1.
Author Contributions
G.Y. contributed to the experimental design, cucumber planting and sampling, tobacco planting and experimenting, data processing and result analysis and writing. Q.C., X.W. and X.M. contributed to analyze the data in experiment. H.F. and N.C. revised the paper. All authors read and approved the final manuscript.
Funding
This study was financially supported by the Nature Science Foundation of Liaoning Province (20170540802) and Key Projects of Basic Scientific Research Projects in Liaoning Province Colleges and Universities (2017004).
Acknowledgments
We thank Zhihong Zhang of Shenyang Agricultural University for donating the carrier.
Conflicts of Interest
The authors declare that this research was conducted in the absence of any financial or commercial relationships that could be interpreted as a potential conflict of interest.
References
- Wang, H.; Li, S.; Guan, W. Identification Technology for Resistance to Target Leaf Spot of Cucumber and Identification for Variety Resistance. China Veg. 2008, 1, 26–27. [Google Scholar]
- Cavatorta, J.; Moriarty, G.; Henning, M.; Glos, M.; Kreitinger, M.; Munger, H.M.; Jahn, M. ‘Marketmore 97’: A Monoecious Slicing Cucumber Inbred with Multiple Disease and Insect Resistances. Hort Sci. 2007, 42, 707–709. [Google Scholar] [CrossRef]
- Wehner, T.C. Gene List 2010 for Cucumber. Cucurbit Genet. Coop. Rpt. 2010, 28, 105–141. [Google Scholar]
- Staub, J.E.; Crubaugh, L.K. Cucumber Inbred Line USDA 6632E. Cucurbit Gen. Coop. Rep. 2001, 24, 6–7. [Google Scholar]
- Abul-Hayja, Z.; Williams, P.H.; Peterson, C.E. Inheritance of resistance to anthracnose and target leaf spot in cucumbers. Plant Dis. Rep. 1978, 62, 43–45. [Google Scholar]
- Yang, L.; Koo, D.H.; Li, Y.; Zhang, X.; Luan, F.; Havey, M.J.; Jiang, J.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.; Mao, A.; Dong, C.; Liu, H.; Yu, S.; Guo, Y.D.; Weng, Y.; Xu, Y. Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber, Cucumis sativus L. Theor. Appl. Genet. 2015, 128, 2495–2506. [Google Scholar] [CrossRef] [PubMed]
- Strugala, R.; Delventhal, R.; Schaffrath, U. An organ-specific view on non-host resistance. Front. Plant Sci. 2015, 6, 526. [Google Scholar] [CrossRef] [PubMed]
- Humphry, M.; Consonni, C.; Panstruga, R. Mlo-based powdery mildew immunity: Silver bullet or simply non-host resistance? Mol. Plant Pathol. 2006, 7, 605–610. [Google Scholar] [CrossRef]
- Lipka, U.; Fuchs, R.; Lipka, V. Arabidopsis non-host resistance to powdery mildews. Curr. Opin. Plant Biol. 2008, 11, 404–411. [Google Scholar] [CrossRef]
- Thordal-Christensen, H. Fresh insights into processes of nonhost resistance. Curr. Opin. Plant Biol. 2003, 6, 351–357. [Google Scholar] [CrossRef]
- Nürnberger, T.; Brunner, F. Innate immunity in plants and animals: Emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr. Opin. Plant Biol. 2002, 5, 318–324. [Google Scholar] [CrossRef]
- Panstruga, R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 2005, 33, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Devoto, A.; Hartmann, H.A.; Piffanelli, P.; Elliott, C.; Simmons, C.; Taramino, G.; Goh, C.S.; Cohen, F.E.; Emerson, B.C.; Schulze-Lefert, P. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. J. Mol. Evol. 2003, 56, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Kusch, S.; Pesch, L.; Panstruga, R. Comprehensive phylogenetic analysis sheds light on the diversity and origin of the MLO family of integral membrane proteins. Genome Bio. Evol. 2016, 8, 878–895. [Google Scholar] [CrossRef] [PubMed]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulzelefert, P. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Consonni, C.; Bednarek, P.; Humphry, M.; Francocci, F.; Ferrari, S.; Harzen, A.; Themaat, E.V.L.; Panstruga, R. Tryptophan-derived metabolites are required for antifungal defense in the Arabidopsis mlo2 mutant. Plant Physiol. 2010, 152, 1544–1561. [Google Scholar] [CrossRef] [PubMed]
- Gruner, K.; Zeier, T.; Aretz, C.; Zeier, J. A critical role for Arabidopsis MILDEW RESISTANCE LOCUS O2 in systemic acquired resistance. Plant J. 2018, 94, 1064–1082. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.; Hückelhoven, R.; Beckhove, U.; Nagarajan, S.; Kogel, K.H. A compromised Mlo pathway affects the response of barley to the necrotrophic fungus bipolaris sorokiniana (teleomorph: Cochliobolus sativus) and its toxins. Phytopathology 2001, 91, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Tilmony, R.; He, S.Y. The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 2002, 1, e0039. [Google Scholar] [CrossRef] [PubMed]
- Slusarenko, A.J.; Schlaich, N.L. Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol. Plant Pathol. 2003, 4, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Tines, M.; Choi, Y.J.; Kemen, E.; Ploch, S.; Holub, E.B.; Shin, H.D.; Jones, J.D.G. A new species of Albugo parasitic to Arabidopsis thaliana reveals new evolutionary patterns in white blister rusts (Albuginaceae). Persoonia 2009, 22, 123–128. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.; Herbert, C.; Sreenivasaprasad, S.; Khatib, M.; Esquerré-Tugayé, M.; Dumas, B. A novel Arabidopsis-Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol. Plant-Microbe Interact. 2004, 17, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Garcia, J.; Gruner, K.; Reinstädler, A.; Kemen, A.; Kemen, E.; Cao, L.; Takken, F.L.W.; Reitz, M.U.; Schäfer, P.; O’Connell, R.J.; et al. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 2017, 7, 3913. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, D.; Cui, N.; Yu, Y.; Yu, G.; Fan, H. Transcriptome and miRNA analyses of the response to Corynespora cassiicolain cucumber. Sci. Rep. 2018, 8, 7798. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.C.; Yu, Y.; Fan, H.; Zhang, D.; Cui, N.; Wang, X.; Jia, S.; Yang, Y.; Zhao, J. Analysis of protein regulation in the leaves of resistant cucumber after inoculation with Corynespora cassiicola: A proteomic approach. Biochemistry (Moscow) 2019, 84, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.; Cunningham-Bussel, A. Beyond oxidative stress: An immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 2013, 13, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Miller, G.; Morales, J.; Shulaev, V.; Torres, M.A.; Mittler, R. Respiratory burst oxidases: The engines of ROS signaling. Curr. Opin. Plant Biol. 2011, 14, 691–699. [Google Scholar] [CrossRef]
- Singh, R.; Parihar, P.; Singh, S.; Mishra, R.K.; Singh, V.P.; Prasad, S.M. Reactive oxygen species signaling and stomatal movement: Current updates and future perspectives. Redox Biol. 2017, 11, 213–218. [Google Scholar] [CrossRef]
- Cui, F.; Wu, H.; Safronov, O.; Zhang, P.; Kumar, R.; Kollist, H.; Salojärvi, J.; Panstruga, R.; Overmyer, K. Arabidopsis MLO2 is a negative regulator of sensitivity to extracellular ROS. Plant Cell Environ. 2018, 41, 782–796. [Google Scholar] [CrossRef]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Mori, I.C.; Pei, Z.M.; Leonhardt, N.; Torres, M.A.; Dangl, J.L.; Bloom, R.E.; Bodde, S.; Jones, J.D.; Schroeder, J.I. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003, 22, 2623–2633. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Bio. 2017, 38, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant-pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Suzuki, N.; Miller, G.; Salazar, C.; Mondal, H.A.; Shulaev, E.; Cortes, D.F.; Shuman, J.N.; Luo, X.Z.; Shah, J.; Schlauch, K.; et al. Temporal-Spatial Interaction between Reactive Oxygen Species and Abscisic Acid Regulates Rapid Systemic Acclimation in Plants. Plant Cell 2013, 25, 3553–3569. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Duan, C.; Chen, P.; Li, Q.; Dai, S.; Sun, L.; Ji, K.; Sun, Y.; Xu, W.; et al. The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J. Plant Physiol. 2012, 169, 1874–1882. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; Mclachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of abi3 to execute an aba-dependent growth arrest during germination. Plant J. 2010, 32, 317–328. [Google Scholar] [CrossRef]
- Mahesh, H.M.; Murali, M.; Anup, C.P.M.; Melvin, P.; Sharada, M.S. Salicylic acid seed priming instigates defense mechanism by inducing PR-Proteins in Solanum melongena L. upon infection with Verticillium dahliae Kleb. Plant Physiol. Bioch. 2017, 117, 12–23. [Google Scholar] [CrossRef]
- Soliman, M.H.; Elmohamedy, R.S.R. Induction of Defense-Related Physiological and Antioxidant Enzyme Response against Powdery Mildew Disease in Okra (Abelmoschus esculentus L.) Plant by Using Chitosan and Potassium Salts. Mycobiology 2017, 45, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Analyses of transcriptome and microRNAs in response to cucumber infection by Corynespora cassiicola. Ph.D. Thesis, Shenyang Agricultural University, Shenyang, China, 2018. [Google Scholar]
- Kim, D.S.; Hwang, B.K. The pepper MLO gene, CaMLO2, is involved in the susceptibility cell-death response and bacterial and oomycete proliferation. Plant J. Cell Mol. Bio. 2012, 72, 843–855. [Google Scholar] [CrossRef]
- Panstruga, R. Discovery of novel conserved peptide domains by ortholog comparison within plant multi-protein families. Plant Mol. Biol. 2005, 59, 485–500. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; Van, D.R.; Van, D.L.T.; Diergaarde, P.; Groenendijk, J. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Yu, G.; Wang, X.; Chen, Q.; Cui, N.; Yu, Y.; Fan, H. Cucumber Mildew Resistance Locus O Interacts with Calmodulin and Regulates Plant Cell Death Associated with Plant Immunity. Int. J. Mol. Sci. 2019, 20, 2995. [Google Scholar] [CrossRef] [PubMed]
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored botrytis-induced kinase1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Freialdenhoven, A.; Peterhänsel, C.; Kurth, J.; Kreuzaler, F.; Schulze-Lefert, P. Identification of Genes Required for the Function of Non-Race-Specific mlo Resistance to Powdery Mildew in Barley. Plant Cell 1996, 8, 5–14. [Google Scholar] [CrossRef]
- Chen, Z.; Hartmann, H.A.; Wu, M.J.; Friedman, E.J.; Chen, J.G.; Pulley, M.; Schulze-Lefert, P.; Panstruga, R.; Jones, A.M. Expression analysis of the ATMLO, gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol. Biol. 2006, 60, 583–597. [Google Scholar] [CrossRef]
- Berg, J.A.; Appiano, M.; Martínez, M.S.; Hermans, F.W.K.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015, 15, 243. [Google Scholar] [CrossRef]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W.; et al. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Torres, M.A.; Dangl, J.L.; Jones, J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 2002, 99, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.H.; Lee, S.C. Function of ABA in Stomatal Defense against Biotic and Drought Stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef] [PubMed]
- Fones, H.; Preston, G.M. Reactive oxygen and oxidative stress tolerance in plant pathogenic pseudomonas. FEMS Microbiol. Lett. 2012, 327, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zurbriggen, M.D.; Carrillo, N.; Hajirezaei, M.R. ROS signaling in the hypersensitive response: When, where and what for? Plant Signal. Behav. 2010, 5, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Expl. Bot. 2018, 69, 3465–3476. [Google Scholar] [CrossRef]
- Monshausen, G.B.; Haswell, E.S. A force of nature: Molecular mechanisms of mechanoperception in plants. J. Exp. Bot. 2013, 64, 4663–4680. [Google Scholar] [CrossRef] [PubMed]
- Hafizur, R.; You-Ping, X.; Xuan-Rui, Z.; Xin-Zhong, C. Brassica napus Genome Possesses Extraordinary High Number of CAMTA Genes and CAMTA3 Contributes to PAMP Triggered Immunity and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 581. [Google Scholar]
- Michie, K.; Ikuko, O.; Kazuhito, K.; Naohiko, Y.; Masayuki, F.; Ko, S.; Noriyuki, D.; Hirofumi, Y. Calcium-dependent protein kina-ses regulate the production of reactive oxygen species by potato NADPHoxi-dase. Plant Cell 2007, 19, 1065–1080. [Google Scholar]
- Beffagna, N.; Buffoli, B.; Busi, C. Modulation of reactive oxygen speciesproduction during osmotic stress in Arabidopsis thaliana cultured cells: Involvement of the plasma membrane Ca2+-ATPase and H+-ATPase. Plant Cell Physiol. 2005, 46, 1326–1339. [Google Scholar] [CrossRef]
- Choi, H.W.; Lee, D.H.; Hwang, B.K. The pepper calmodulin gene CaCaM1 is involved in reactive oxygen species and nitric oxide generation required for cell death and the defense response. Mol. Plant-Microbe Interactions 2009, 22, 1389–1400. [Google Scholar] [CrossRef]
- Abuqamar, S.; Moustafa, K.; Tran, L.P. Mechanisms and strategies of plant defense against Botrytis cinerea. Crit. Rev. Biotechnol. 2017, 61, 1–16. [Google Scholar]
- Xue, M.; Yi, H. Enhanced Arabidopsis disease resistance against Botrytis cinerea induced by sulfur dioxide. Ecotoxicol. Environ. Saf. 2017, 147, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Huffaker, A.; Ryan, C.A. Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 2007, 104, 10732–10736. [Google Scholar] [CrossRef]
- Lim, C.W.; Lee, S.C. Functional roles of the pepper MLO protein gene, CaMLO2, in abscisic acid signaling and drought sensitivity. Plant Mol Biol. 2013, 85, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vallet, A.; López, G.; Ramos, B.; Delgado-Cerezo, M.; Riviere, M.P.; Llorente, F.; Fernández, P.V.; Miedes, E.; Estevez, J.M.; Grant, M.; et al. Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol. 2012, 160, 2109–2124. [Google Scholar] [CrossRef] [PubMed]
- Ulferts, S.; Delventhal, R.; Splivallo, R.; Karlovsky, P.; Schaffrath, U. Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol. 2015, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Sivakumaran, A.; Akinyemi, A.; Mandon, J.; Cristescu, S.M.; Hall, M.A.; Harren, F.J.; Mur, L.A. ABA suppresses Botrytis cinerea Elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 2016, 7, 709. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q. Analysis of CsTCTP regulated signal pathway and construction of yeast two-hybrid system in cucumber. Master’s Thesis, Shenyang Agricultural University, Shenyang, China, 2017. [Google Scholar]
- Jiang, M.; Zhang, J. Cross-talk between calcium and reactive oxygen species originated from NADPH oxidase in abscisic acid-induced antioxidant de-fence in leaves of maize seedlings. Plant Cell Environ. 2003, 26, 929–939. [Google Scholar] [CrossRef]
- Xu, S. Abscisic acid activates a Ca2+-calmodulin-stimulated protein kinase involved in antioxidant defense in maize leaves. Acta Biochim. Biophys. Sin. 2010, 42, 646–655. [Google Scholar] [CrossRef][Green Version]
- Srinivas, A.; Raghavendra, A.S. Convergence and Divergence of Signaling Events in Guard Cells during Stomatal Closure by Plant Hormones or Microbial Elicitors. Front. Plant Sci. 2016, 7. [Google Scholar]
- Parvin, S.; Lee, O.R.; Sathiyaraj, G.; Khorolragchaa, A.; Kim, Y.J.; Devi, B.S.R.; Yang, D.C. Interrelationship between calmodulin (CaM) and H2O2 in abscisic acid-induced antioxidant defense in the seedlings of Panax ginseng. Mol. Biol. Rep. 2012, 39, 7327–7338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKYW46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Bioch. 2016, 108, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Ye, T.; Chen, F.; Cheng, Z.; Wang, Y.; Yang, P.; Zhang, Y.; Cha, Z. Manipulation of arginase expression modulates abiotic stress tolerance in Arabidopsis: Effect on arginine metabolism and ROS accumulation. J. Exp. Bot. 2013, 64, 1367–1379. [Google Scholar] [CrossRef]
- Zhong, R.; Kays, S.J.; Schroeder, B.P.; Ye, Z.H. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 2002, 14, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, J.; Chen, X.; Gao, Z.; Xuan, W.; Xu, S.; Ding, X.; Shen, W. Carbon monoxide alleviatescadmium-induced oxidative damage by modulating glutathione metabolism in the roots of Medicago sativa. New Phytol. 2008, 177, 155–166. [Google Scholar] [PubMed]
- Huang, B.K.; Xu, S.; Xuan, W.; Li, M.; Cao, Z.Y.; Liu, K.L.; Ling, T.F.; Shen, W.B. Carbon monoxide alleviates salt-induced oxidative damage in wheat seedling leaves. J. Integr. Plant Biol. 2006, 48, 249–254. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).