Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean
Abstract
:1. Introduction
2. Results
2.1. Physico-Chemical Properties of GmCAMTA Proteins
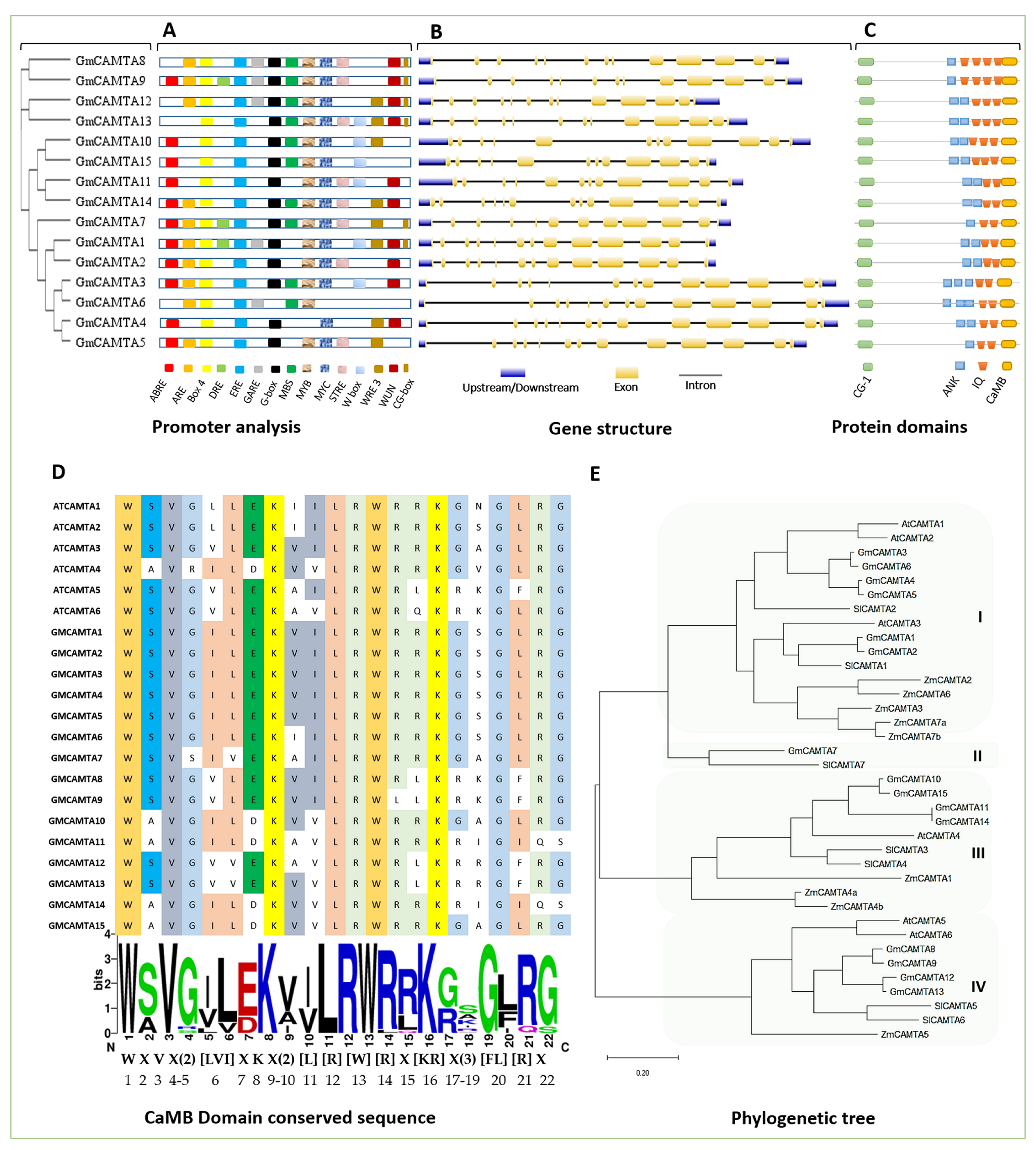
2.2. Phylogenetics and Structure of GmCAMTAs
2.3. miRNA Targets in GmCAMTA Transcripts
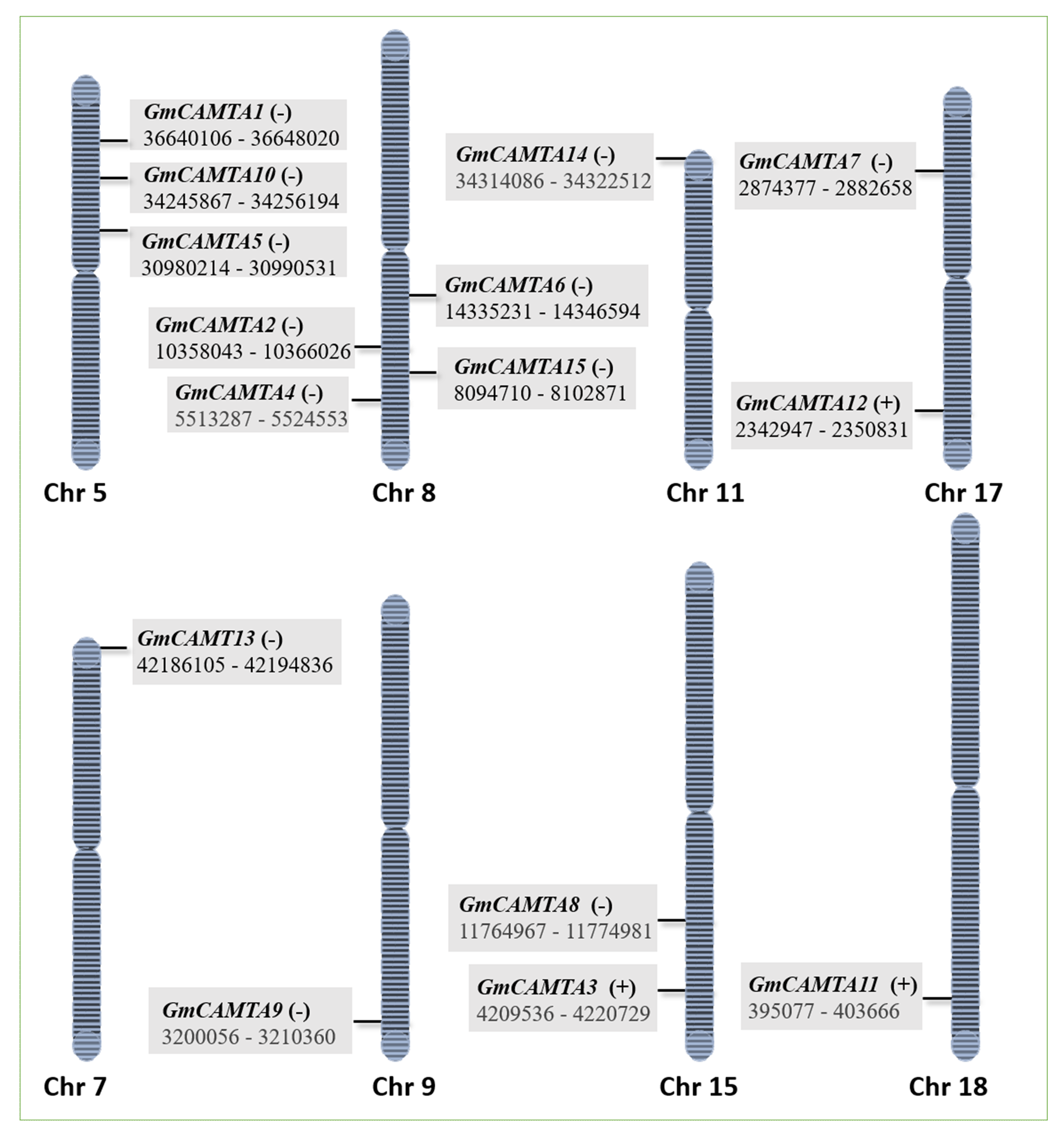
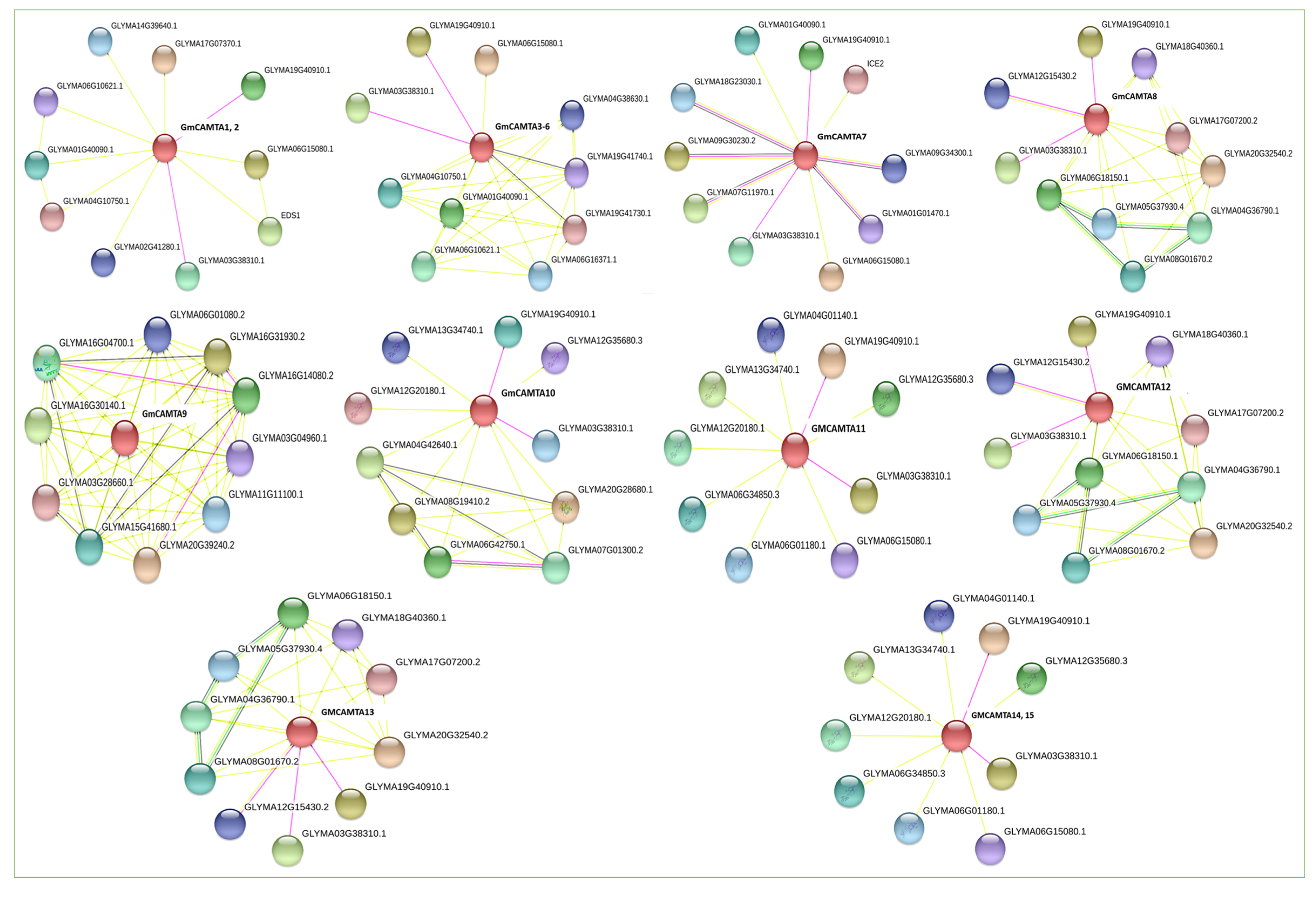
2.4. Chromosomal Distribution and Regulatory Network of GmCAMTAs
2.5. GmCAMTAs as Early Drought Stress-Responsive TFs
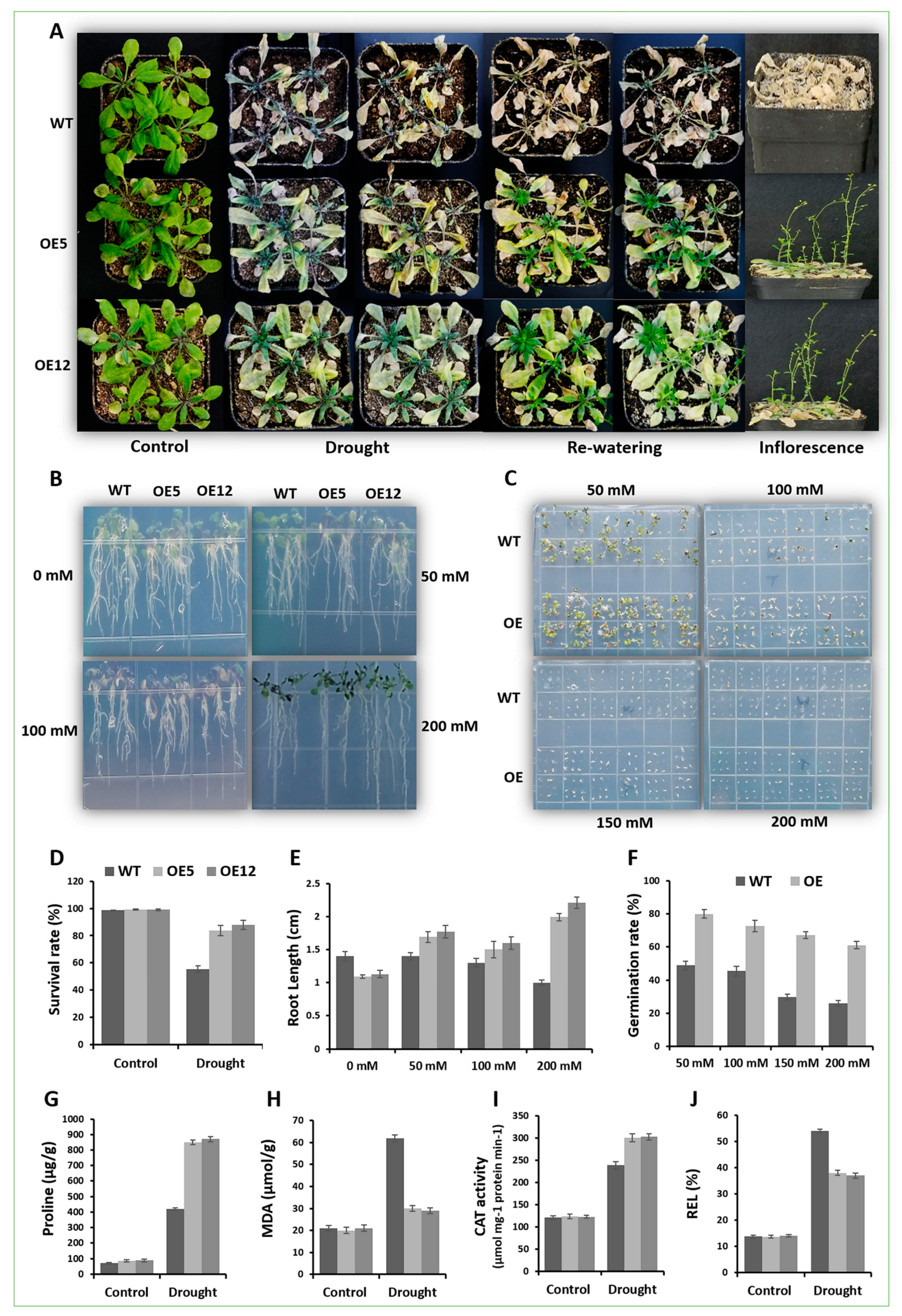
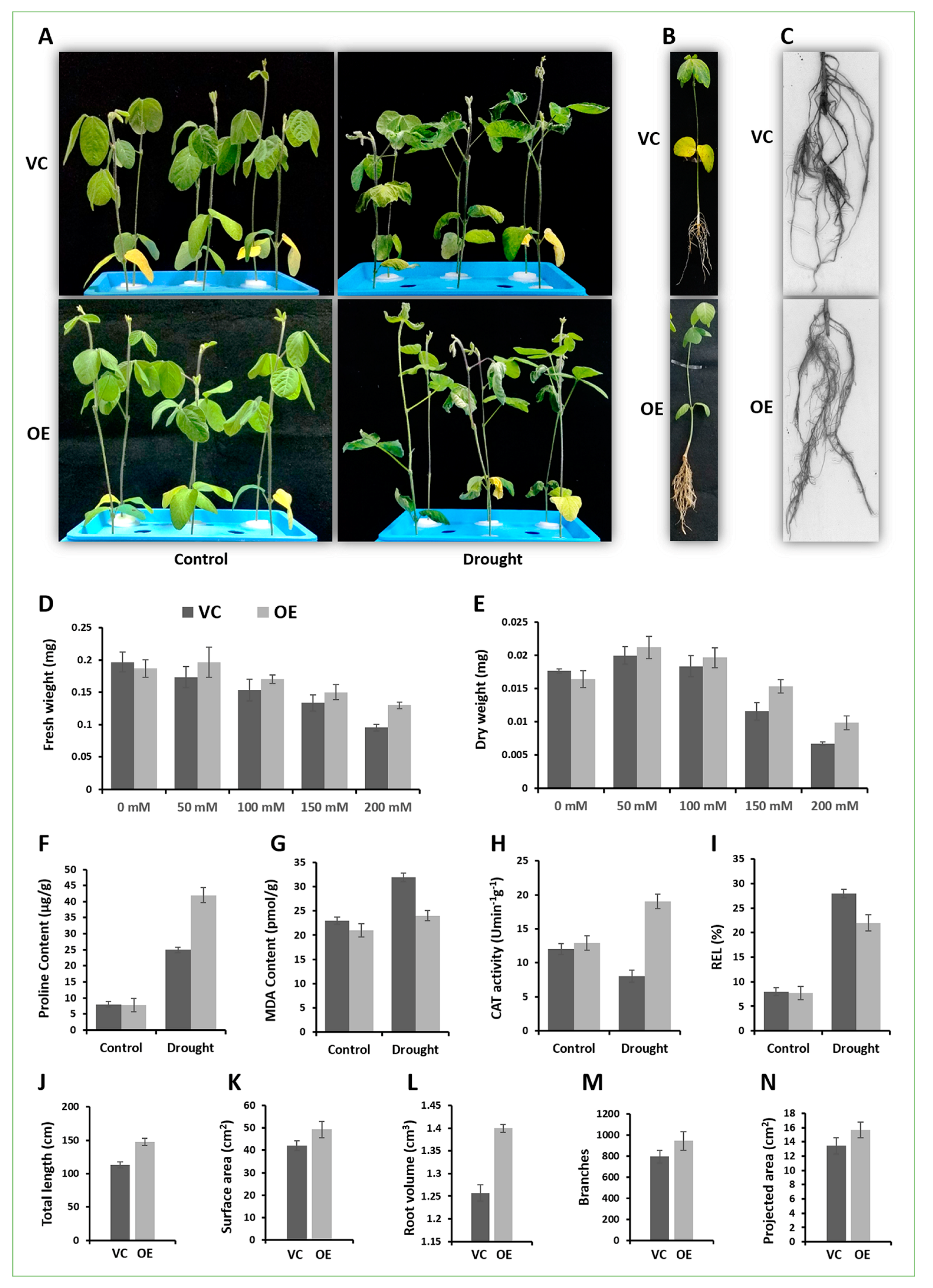
2.6. Arabidopsis Overexpressing GmCAMTA12 Exhibited Enhanced Drought Tolerance
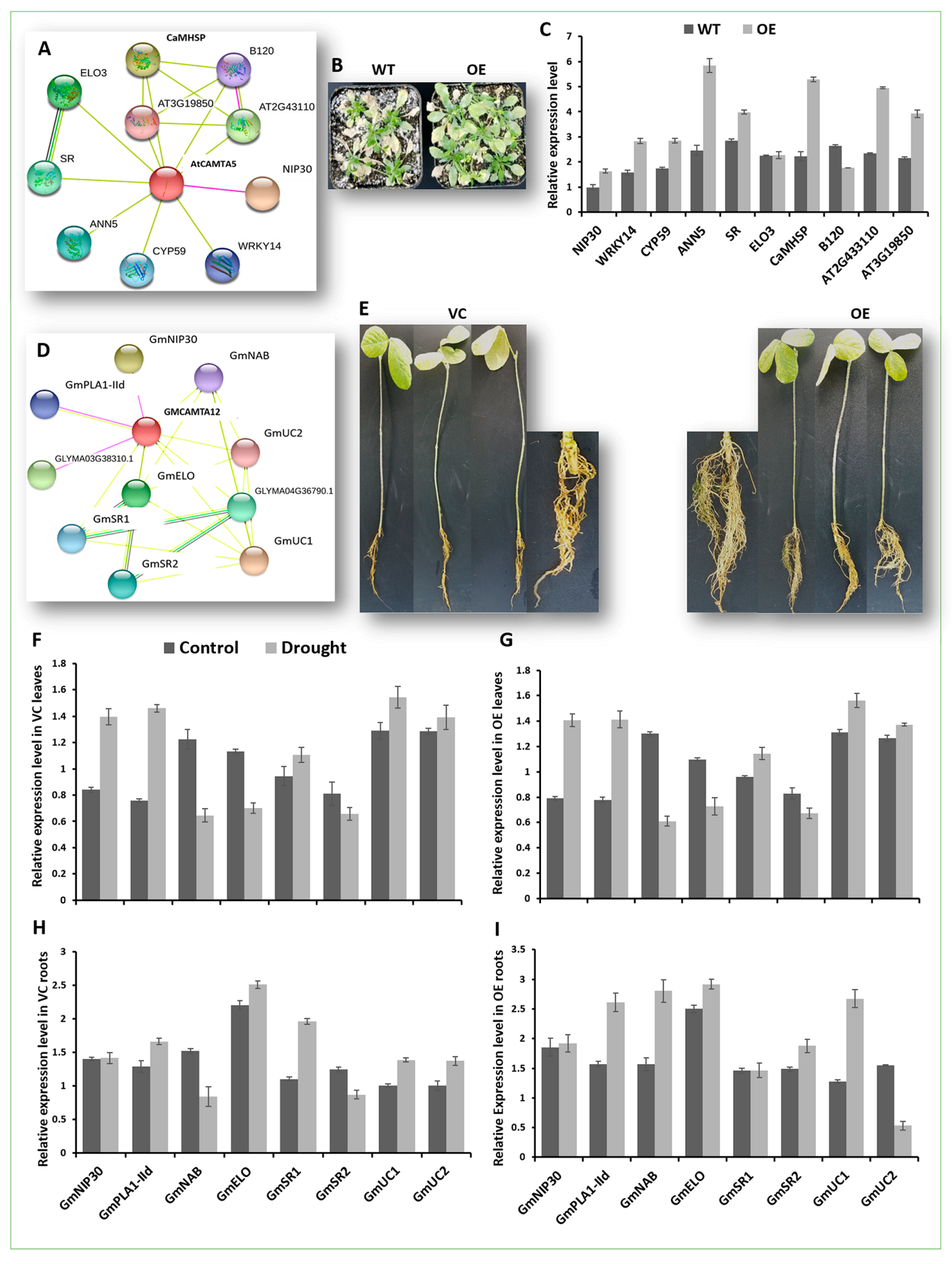
2.7. GmCAMTA12 Overexpression Regenerated More Developed and Drought-Efficient Hairy Roots in Soybean
2.8. Expression Analysis of GmCAMTA12 Orthologues’ Regulatory Network in Arabidopsis
2.9. GmCAMTA12 Overexpression Orchestrated Downstream Genes in Transgenic Hairy Roots
3. Discussion
4. Materials and Methods
4.1. In Silico Analysis
4.2. Expression Analysis
4.3. Gene Transformation and Drought Assays
4.4. Expression Analysis of the GmCAMTA12 Orthologue’s Regulatory Network in wt and OE Arabidopsis
4.5. Analysis of GmCAMTA12 Regulatory Network in Chimeric Soybean Plants
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CAMTA | Calmodulin Binding Transcription Activator |
| CaM | Calmodulin (Calcium-Modulated Protein) |
| CML | Calmodulin-Like |
| CBL | Calcineurin B-like |
| ML | Maximum Likelihood |
| OE | Overexpression |
| PEG | Poly Ethylene Glycol |
| TF | Transcription Factor |
| VC | Vector Control |
| WT | Wild Type |
References
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M. CToulmin, Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinozaki, K.; Uemura, M.; Bailey-Serres, J.; Bray, E. Responses to abiotic stress. In Biochemistry and Molecular Biology of Plants, 2nd ed.; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; Wiley: Oxford, UK, 2015; pp. 1051–1100. [Google Scholar]
- Prudhomme, C.; Giuntoli, I.; Robinson, E.L.; Clark, D.B.; Arnell, N.W.; Dankers, R.; Fekete, B.M.; Franssen, W.; Gerten, D.; Gosling, S.N.; et al. DWisser, Hydrological droughts in the 21 st century, hotspots and uncertainties from a global multimodel ensemble experiment. Proc. Natl. Acad. Sci. USA 2014, 111, 3262–3267. [Google Scholar] [CrossRef]
- Wei, W.; Liang, D.-W.; Bian, X.-H.; Shen, M.; Xiao, J.-H.; Zhang, W.-K.; Ma, B.; Lin, Q.; Lv, J.; Chen, X.; et al. GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Y.; Zhu, J.-K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Li, Y.; Chen, D.; Liu, H.; Zhu, D.; Xiang, Y. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis). Sci. Rep. 2016, 6, 24520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.-A.; Li, M.-Z.; Wang, S.-M.; Yin, H.-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Chung, W.S.; Yun, D.-J.; Cho, M.J. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2009, 2, 13–21. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Reddy, V.S.; Golovkin, M. A Calmodulin Binding Protein from Arabidopsis Is Induced by Ethylene and Contains a DNA-Binding Motif. Biochem. Biophys. Res. Commun. 2000, 279, 762–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Poovaiah, B.W. A Calmodulin-binding/CGCG Box DNA-binding Protein Family Involved in Multiple Signaling Pathways in Plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouché, N.; Scharlat, A.; Snedden, W.; Bouchez, D.; Fromm, H. A Novel Family of Calmodulin-binding Transcription Activators in Multicellular Organisms. J. Biol. Chem. 2002, 277, 21851–21861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zegzouti, H.; Jones, B.; Frasse, P.; Marty, C.; Maitre, B.; Latché, A.; Pech, J.-C.; Bouzayen, M. Ethylene-regulated gene expression in tomato fruit: Characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. Plant J. 1999, 18, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, T.; Xu, L.; Pi, E.; Wang, S.; Wang, H.; Shen, C. Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Front. Plant Sci. 2015, 6, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Pan, X.; Ge, T.; Yi, S.; Lv, Q.; Zheng, Y.; Ma, Y.; Liu, X.; Xie, R. Genome-wide identification of citrus CAMTA genes and their expression analysis under stress and hormone treatments. J. Hortic. Sci. Biotechnol. 2019, 94, 331–340. [Google Scholar] [CrossRef]
- Wei, M.; Xu, X.; Li, C. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci. Rep. 2017, 7, 17910. [Google Scholar] [CrossRef] [Green Version]
- Kakar, K.U.; Nawaz, Z.; Cui, Z.; Cao, P.; Jin, J.; Shu, Q.; Ren, X. Evolutionary and expression analysis of CAMTA gene family in Nicotiana tabacum yielded insights into their origin, expansion and stress responses. Sci. Rep. 2018, 8, 10322. [Google Scholar] [CrossRef]
- Meer, L.; Mumtaz, S.; Labbo, A.M.; Khan, M.J.; Sadiq, I. Genome-wide identification and expression analysis of calmodulin-binding transcription activator genes in banana under drought stress. Sci. Hortic. 2019, 244, 10–14. [Google Scholar] [CrossRef]
- Büyük, İ.; İlhan, E.; Şener, D.; Özsoy, A.U.; Aras, S. Genome-wide identification of CAMTA gene family members in Phaseolus vulgaris L. and their expression profiling during salt stress. Mol. Biol. Rep. 2019, 46, 2721–2732. [Google Scholar] [CrossRef]
- Wang, G.; Zeng, H.; Hu, X.; Zhu, Y.; Chen, Y.; Shen, C.; Wang, H.; Poovaiah, B.W.; Du, L. Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil. 2015, 386, 205–221. [Google Scholar] [CrossRef]
- Galon, Y.; Aloni, R.; Nachmias, D.; Snir, O.; Feldmesser, E.; Scrase-Field, S.; Boyce, J.M.; Bouché, N.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 2010, 232, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Yang, J.; Xu, Y.-P.; Munyampundu, J.-P.; Cai, X.-Z. Phylogeny of Plant CAMTAs and Role of AtCAMTAs in Nonhost Resistance to Xanthomonas oryzae pv. oryzae. Front. Plant Sci. 2016, 7, 177. [Google Scholar] [CrossRef] [PubMed]
- Rahman, H.; Xu, Y.-P.; Zhang, X.-R.; Cai, X.-Z. Brassica napus Genome Possesses Extraordinary High Number of CAMTA Genes and CAMTA3 Contributes to PAMP Triggered Immunity and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 2016, 7, 581. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wei, F.; Zhou, H.; Liu, N.; Niu, X.; Yan, C.; Zhang, L.; Han, S.; Hou, C.; Wang, D. TaCAMTA4, a Calmodulin-Interacting Protein, Involved in Defense Response of Wheat to Puccinia triticina. Sci. Rep. 2019, 9, 641. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Poovaiah, B.W. An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 2000, 275, 38467–38473. [Google Scholar] [CrossRef] [PubMed]
- [Benn, G.; Wang, C.-Q.; Hicks, D.R.; Stein, J.; Guthrie, C.; Dehesh, K. A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J. 2014, 80, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Peng, H.; Whitaker, B.D.; Jurick, W.M. Differential expression of calcium/calmodulin-regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiol. Plant. 2013, 148, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.N.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154. [Google Scholar] [CrossRef]
- Nie, H.; Zhao, C.; Wu, G.; Wu, Y.; Chen, Y.; Tang, D. SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 2012, 158, 1847–1859. [Google Scholar] [CrossRef]
- Pandey, N.; Ranjan, A.; Pant, P.; Tripathi, R.K.; Ateek, F.; Pandey, H.P.; Patre, U.V.; Sawant, S.V. CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genom. 2013, 14, 216. [Google Scholar] [CrossRef]
- Doherty, C.J.; van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013, 75, 364–376. [Google Scholar] [CrossRef]
- Prasad, K.V.S.K.; Abdel-Hameed, A.A.E.; Xing, D.; Reddy, A.S.N. Global gene expression analysis using RNA-seq uncovered a new role for SR1/CAMTA3 transcription factor in salt stress. Sci. Rep. 2016, 6, 27021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; An, C.; Park, S.; Gilmour, S.J.; Wang, L.; Renna, L.; Brandizzi, F.; Grumet, R.; Thomashow, M.F. CAMTA-Mediated Regulation of Salicylic Acid Immunity Pathway Genes in Arabidopsis Exposed to Low Temperature and Pathogen Infection. Plant Cell 2017, 29, 2465–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shkolnik, D.; Finkler, A.; Pasmanik-Chor, M.; Fromm, H. CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: A Key Regulator of Na+ Homeostasis during Germination. Plant Physiol. 2019, 180, 1101–1118. [Google Scholar] [CrossRef] [PubMed]
- 2019 Soystats. Available online: http://soystats.com/wp-content/uploads/2019-SoyStats-Web.pdf (accessed on 10 August 2019).
- Li, Y.; Chen, Q.; Nan, H.; Li, X.; Lu, S.; Zhao, X.; Liu, B.; Guo, C.; Kong, F.; Cao, D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS ONE 2017, 12, e0179554. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.S.; Kim, M.C.; Yoo, J.H.; Moon, B.C.; Koo, S.C.; Park, B.O.; Lee, J.H.; Koo, Y.D.; Han, H.J.; Lee, S.Y.; et al. Isolation of a Calmodulin-binding Transcription Factor from Rice (Oryza sativa L.). J. Biol. Chem. 2005, 280, 40820–40831. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Gong, P.; Reddig, K.; Mitra, M.; Guo, P.; Li, H.-S. The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell 2006, 127, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Finkler, A.; Kaplan, B.; Fromm, H. Ca-Responsive cis-Elements in Plants. Plant Signal. Behav. 2007, 2, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.H.; Mostofa, M.G.; Li, W.; van Ha, C.; Watanabe, Y.; Le, D.T.; Thao, N.P.; Tran, L.-S.P. The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ. Exp. Bot. 2018, 151, 12–20. [Google Scholar] [CrossRef]
- Shi, W.-Y.; Du, Y.-T.; Ma, J.; Min, D.-H.; Jin, L.-G.; Chen, J.; Chen, M.; Zhou, Y.-B.; Ma, Y.-Z.; Xu, Z.-S.; et al. The WRKY Transcription Factor GmWRKY12 Confers Drought and Salt Tolerance in Soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Xia, Z.; Xu, Z.; Wei, Y.; Wang, M. Overexpression of the Maize Sulfite Oxidase Increases Sulfate and GSH Levels and Enhances Drought Tolerance in Transgenic Tobacco. Front. Plant Sci. 2018, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, K.; Zhang, J.; Li, X.; Xu, K.; Zhang, Y.; Qi, J.; Yu, D.; Wang, J.; Li, C. JcDREB2, a Physic Nut AP2/ERF Gene, Alters Plant Growth and Salinity Stress Responses in Transgenic Rice. Front. Plant Sci. 2017, 8, 306. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-induced electrolyte leakage: The role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.T.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noman, M.; Jameel, A.; Qiang, W.-D.; Ahmad, N.; Liu, W.-C.; Wang, F.-W.; Li, H.-Y. Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean. Int. J. Mol. Sci. 2019, 20, 4849. https://doi.org/10.3390/ijms20194849
Noman M, Jameel A, Qiang W-D, Ahmad N, Liu W-C, Wang F-W, Li H-Y. Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean. International Journal of Molecular Sciences. 2019; 20(19):4849. https://doi.org/10.3390/ijms20194849
Chicago/Turabian StyleNoman, Muhammad, Aysha Jameel, Wei-Dong Qiang, Naveed Ahmad, Wei-Can Liu, Fa-Wei Wang, and Hai-Yan Li. 2019. "Overexpression of GmCAMTA12 Enhanced Drought Tolerance in Arabidopsis and Soybean" International Journal of Molecular Sciences 20, no. 19: 4849. https://doi.org/10.3390/ijms20194849





