Cellular and Molecular Mechanisms Mediated by recPrPC Involved in the Neuronal Differentiation Process of Mesenchymal Stem Cells
Abstract
:1. Introduction
2. Results
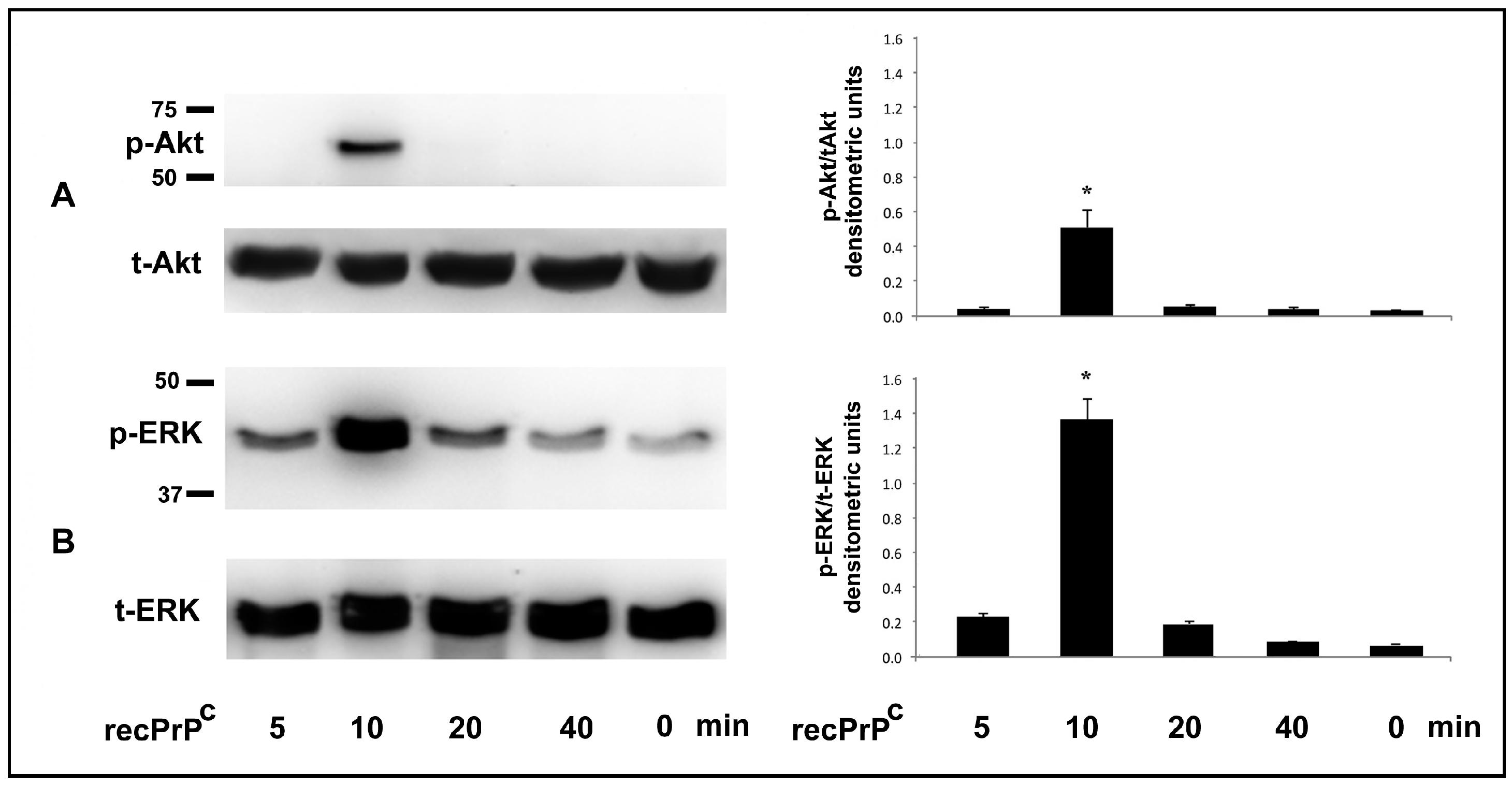
2.1. Role of recPrPC in Signal Pathways of hDPSCs
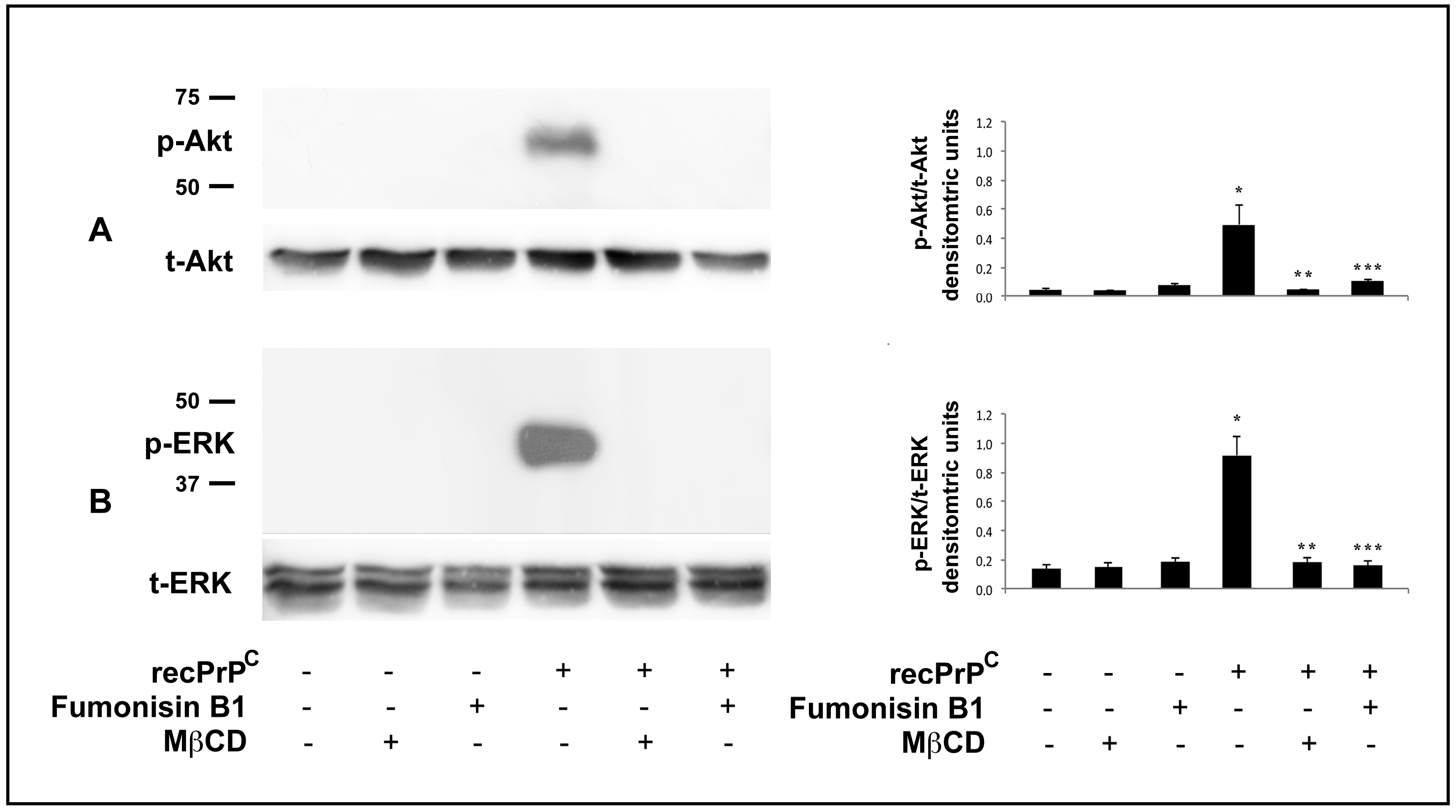
2.2. Role of Endogenous PrPC in the Modulation of Cell Signaling Induced by recPrPC
2.3. Role of Lipid Rafts in the Modulation of Cell Signaling Induced by recPrPC
2.4. Role of recPrPC in the Neuronal Differentiation of hDPSCs
2.5. Role of Endogenous PrPC in the Neuronal Differentiation Process Induced by recPrPC
3. Discussion
4. Materials and Methods
4.1. Research Ethics
4.2. Isolation of Stem Cells Derived from Human Dental Pulp
4.3. Treatments
4.4. Knockdown PrPC by siRNA
4.5. Western Blot Analysis
4.6. Flow Cytometry Analysis
4.7. Immunofluorescence Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, D.R.; Qin, K.; Herms, J.W.; Madlung, A.; Manson, J.; Strome, R.; Fraser, P.E.; Kruck, T.; von Bohlen, A.; Schulz-Schaeffer, W.; et al. The cellular prion protein binds copper in vivo. Nature 1997, 390, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Mattei, V.; Martellucci, S.; Santilli, F.; Manganelli, V.; Garofalo, T.; Candelise, N.; Caruso, A.; Sorice, M.; Scaccianoce, S.; Misasi, R. Morphine Withdrawal Modifies Prion Protein Expression in Rat Hippocampus. PLoS ONE 2017, 12, e0169571. [Google Scholar] [CrossRef] [PubMed]
- Lewis, V.; Hooper, N.M. The role of lipid rafts in prion protein biology. Front Biosci. 2011, 16, 151–168. [Google Scholar] [CrossRef]
- Sorice, M.; Mattei, V.; Tasciotti, V.; Manganelli, V.; Garofalo, T.; Misasi, R. Trafficking of PrPC to mitochondrial raft-like microdomains during cell apoptosis. Prion 2012, 6, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Prusiner, S.B.; Scott, M.R.; DeArmond, S.J.; Cohen, F.E. Prion protein biology. Cell 1998, 93, 337–348. [Google Scholar] [CrossRef]
- Parizek, P.; Roeckl, C.; Weber, J.; Flechsig, E.; Aguzzi, A.; Raeber, A.J. Similar turnover and shedding of the cellular prion protein in primary lymphoid and neuronal cells. J. Biol. Chem. 2001, 276, 44627–44632. [Google Scholar] [CrossRef]
- Mattei, V.; Garofalo, T.; Misasi, R.; Circella, A.; Manganelli, V.; Lucania, G.; Pavan, A.; Sorice, M. Prion protein is a component of the multimolecular signaling complex involved in T cell activation. FEBS Lett. 2004, 560, 14–18. [Google Scholar] [CrossRef] [Green Version]
- Mouillet-Richard, S.; Ermonval, M.; Chebassier, C.; Laplanche, J.L.; Lehmann, S.; Launay, J.M.; Kellermann, O. Signal transduction through prion protein. Science 2000, 289, 1925–1928. [Google Scholar] [CrossRef]
- Toni, M.; Spisni, E.; Griffoni, C.; Santi, S.; Riccio, M.; Lenaz, P.; Tomasi, V. Cellular prion protein and caveolin-1 interaction in a neuronal cell line precedes Fyn/Erk 1/2 signal transduction. J. Biomed. Biotechnol. 2006, 2006, 69469. [Google Scholar] [CrossRef]
- Llorens, F.; Carulla, P.; Villa, A.; Torres, J.M.; Fortes, P.; Ferrer, I.; del Río, J.A. PrP(C) regulates epidermal growth factor receptor function and cell shape dynamics in Neuro2a cells. J. Neurochem. 2013, 127, 124–138. [Google Scholar]
- Hirsch, T.Z.; Martin-Lannerée, S.; Mouillet-Richard, S. Functions of the Prion Protein. Prog. Mol. Biol. Transl. Sci. 2017, 150, 1–34. [Google Scholar] [PubMed]
- Hu, W.; Kieseier, B.; Frohman, E.; Eagar, T.N.; Rosenberg, R.N.; Hartung, H.P.; Stüve, O. Prion proteins: Physiological functions and role in neurological disorders. J. Neurol. Sci. 2008, 264, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mattei, V.; Matarrese, P.; Garofalo, T.; Tinari, A.; Gambardella, L.; Ciarlo, L.; Manganelli, V.; Tasciotti, V.; Misasi, R.; Malorni, W.; et al. Recruitment of cellular prion protein to mitochondrial raft-like microdomains contributes to apoptosis execution. Mol. Biol. Cell 2011, 22, 4842–4853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garofalo, T.; Manganelli, V.; Grasso, M.; Mattei, V.; Ferri, A.; Misasi, R.; Sorice, M. Role of mitochondrial raft-like microdomains in the regulation of cell apoptosis. Apoptosis 2015, 20, 621–634. [Google Scholar] [CrossRef]
- Wulf, M.A.; Senatore, A.; Aguzzi, A. The biological function of the cellular prion protein: An update. BMC Biol. 2017, 15, 34. [Google Scholar] [CrossRef]
- Linden, R. The Biological Function of the Prion Protein: A Cell Surface Scaffold of Signaling Modules. Front Mol. Neurosci. 2017, 10, 77. [Google Scholar] [CrossRef]
- Kanaani, J.; Prusiner, S.B.; Diacovo, J.; Baekkeskov, S.; Legname, G. Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J. Neurochem. 2005, 95, 1373–1386. [Google Scholar] [CrossRef] [Green Version]
- Steele, A.D.; Emsley, J.G.; Ozdinler, P.H.; Lindquist, S.; Macklis, J.D. Prion protein (PrPc) positively regulates neural precursor proliferation during developmental and adult mammalian neurogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 3416–3421. [Google Scholar] [CrossRef] [Green Version]
- Miranda, A.; Ramos-Ibeas, P.; Pericuesta, E.; Ramirez, M.A.; Gutierrez-Adan, A. The role of prion protein in stem cell regulation. Reproduction 2013, 146, R91–R99. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Baskokov, I.V. The cellular form of the prion protein guides the differentiation of human embryonic stem cell into neuron-, oligodendrocyte- and astrocyte-committed lineages. Prion 2014, 8, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Martellucci, S.; Manganelli, V.; Santacroce, C.; Santilli, F.; Piccoli, L.; Sorice, M.; Mattei, V. Role of Prion protein-EGFR multimolecular complex during neuronal differentiation of human dental pulp-derived stem cells. Prion 2018, 12, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Mediano, D.R.; Sanz-Rubio, D.; Ranera, R.; Bolea, I.; Martin-Burriel, I. The potential of mesenchymal stem cell in prion research. Zoonoses Public Health 2015, 62, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem cell properties of human dental pulp stem cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Arthur, A.; Rychkov, G.; Shi, S.; Koblar, S.A.; Gronthos, S. Adult human dental pulp stem cells differentiate toward functionally active neurons under appropriate environmental cues. Stem Cells 2008, 7, 1787–1795. [Google Scholar] [CrossRef]
- Suchanek, J.; Soukup, T.; Visek, B.; Ivancakova, R.; Kucerova, L.; Mokry, J. Dental pulp stem cells and their characterization. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc. Czech Repub. 2009, 153, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Koyama, N.; Okubo, Y.; Nakao, K.; Bessho, K. Evaluation of pluripotency in human dental pulp cells. J. Oral Maxillofac Surg. 2009, 67, 501–506. [Google Scholar] [CrossRef]
- Lee, S.H.; Ryu, J.S.; Lee, J.W.; Kwak, D.H.; Ko, K.; Choo, Y.K. Comparison of ganglioside expression between human adipose- and dental pulp-derived stem cell differentiation into osteoblasts. Arch. Pharm. Res. 2010, 33, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Bieberich, E. It’s a lipid’s world: Bioactive lipid metabolism and signaling in neural stem cell differentiation. Neurochem. Res. 2012, 37, 1208–1229. [Google Scholar] [CrossRef]
- Atari, M.; Gil-Recio, C.; Fabregat, M.; García-Fernández, D.; Barajas, M.; Carrasco, M.A.; Jung, H.S.; Alfaro, F.H.; Casals, N.; Prosper, F.; et al. Dental pulp of the third molar: A new source of pluripotent-like stem cells. J. Cell Sci. 2012, 125, 3343–3356. [Google Scholar] [CrossRef]
- Young, F.I.; Telezhkin, V.; Youde, S.J.; Langley, M.S.; Stack, M.; Kemp, P.J.; Waddington, R.J.; Sloan, A.J.; Song, B. Clonal heterogeneity in the neuronal and glial differentiation of dental pulp stem/progenitor cells. Stem Cells Int. 2016, 2016. [Google Scholar] [CrossRef]
- Ullah, I.; Subbarao, R.B.; Kim, E.J.; Bharti, D.; Jang, S.J.; Park, J.S.; Shivakumar, S.B.; Lee, S.L.; Kang, D.; Byun, J.H.; et al. In vitro comparative analysis of human dental stem cells from a single donor and its neurodifferentiation potential evaluated by electrophysiology. Life Sci. 2016, 154, 39–51. [Google Scholar] [CrossRef]
- Chun, S.Y.; Soker, S.; Jang, Y.J.; Kwon, T.G.; Yoo, E.S. Differentiation of human dental pulp stem cells into dopaminergic neuron-like cells in vitro. J. Korean Med. Sci. 2016, 31, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, E.; Koh, S.E.; Maeng, S.; Lee, W.D.; Lim, J.; Shim, I.; Lee, Y.J. Dopaminergic differentiation of neural progenitors derived from placental mesenchymal stem cells in the brains of Parkinson’s disease model rats and alleviation of asymmetric rotational behaviour. Brain Res. 2012, 1466, 158–166. [Google Scholar] [CrossRef]
- Nesti, C.; Pardini, S.; Barachini, S.; D’Alessandro, D.; Siciliano, G.; Murri, L.; Petrini, M.; Vaglini, F. Human dental pulp stem cells protect mouse dopaminergic neurons against MPP+ orrotenone. Brain Res. 2011, 1367, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Harris, D.A.; Huber, M.T.; van Dijken, P.; Shyng, S.L.; Chait, B.T.; Wang, R. Processing of a cellular prion protein: Identification of N- and C-terminal cleavage sites. Biochemistry 1993, 32, 1009–1016. [Google Scholar] [CrossRef]
- Watt, N.T.; Taylor, D.R.; Gillott, A.; Thomas, D.A.; Perera, W.S.; Hooper, N.M. Reactive oxygen species-mediated beta-cleavage of the prion protein in the cellular response to oxidative stress. J. Biol. Chem. 2005, 280, 35914–35921. [Google Scholar] [CrossRef] [PubMed]
- Altmeppen, H.C.; Prox, J.; Puig, B.; Dohler, F.; Falker, C.; Krasemann, S.; Glatzel, M. Roles of endoproteolytic α-cleavage and shedding of the prion protein in neurodegeneration. FEBS J. 2013, 280, 4338–4347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, A.J.; Dibble, J.P.; Evans, E.G.; Millhauser, G.L. A new paradigm for enzymatic control of α-cleavage and β-cleavage of the prion protein. J. Biol. Chem. 2014, 289, 803–813. [Google Scholar] [CrossRef]
- Altmeppen, H.C.; Puig, B.; Dohler, F.; Thurm, D.K.; Falker, C.; Krasemann, S.; Glatzel, M. Proteolytic processing of the prion protein in health and disease. Am. J. Neurodegener Dis. 2012, 1, 15–31. [Google Scholar]
- Borchelt, D.R.; Rogers, M.; Stahl, N.; Telling, G.; Prusiner, S.B. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology 1993, 3, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stahl, N.; Borchelt, D.R.; Prusiner, S.B. Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry 1990, 29, 5405–5412. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.R.; Parkin, E.T.; Cocklin, S.L.; Ault, J.R.; Ashcroft, A.E.; Turner, A.J.; Hooper, N.M. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J. Biol. Chem. 2009, 284, 22590–22600. [Google Scholar] [CrossRef] [PubMed]
- Linsenmeier, L.; Altmeppen, H.C.; Wetzel, S.; Mohammadi, B.; Saftig, P.; Glatzel, M. Diverse functions of the prion protein—Does proteolytic processing hold the key? Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2128–2137. [Google Scholar] [CrossRef]
- Tagliavini, F.; Prelli, F.; Porro, M.; Salmona, M.; Bugiani, O.; Frangione, B. A soluble form of prion protein in human cerebrospinal fluid: Implications for prion-related encephalopathies. Biochem. Biophys. Res. Commun. 1992, 184, 1398–1404. [Google Scholar] [CrossRef]
- Perini, F.; Vidal, R.; Ghetti, B.; Tagliavini, F.; Frangione, B.; Prelli, F. PrP27-30 is a normal soluble prion protein fragment released by human platelets. Biochem. Biophys. Res. Commun. 1996, 223, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.C.; Steele, A.D.; Lindquist, S.; Lodish, H.F. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc. Natl. Acad. Sci. USA 2006, 103, 2184–2189. [Google Scholar] [CrossRef] [Green Version]
- Rigter, A.; Langeveld, J.P.M.; Zijderveld, F.G.V.; Bossers, A. Prion protein self-interactions: A gateway to novel therapeutic strategies? Vaccine 2010, 28, 7810–7823. [Google Scholar] [CrossRef] [Green Version]
- Mattei, V.; Santacroce, C.; Tasciotti, V.; Martellucci, S.; Santilli, F.; Manganelli, V.; Piccoli, L.; Misasi, R.; Sorice, M.; Garofalo, T. Role of lipid rafts in neuronal differentiation of dental pulp-derived stem cells. Exp. Cell Res. 2015, 339, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Martellucci, S.; Santacroce, C.; Manganelli, V.; Santilli, F.; Piccoli, L.; Cassetta, M.; Misasi, R.; Sorice, M.; Mattei, V. Isolation, Propagation and Prion Protein Expression During Neuronal Differentiation Process of Human Dental Pulp Stem Cells. J. Vis. Exp. in press.



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martellucci, S.; Santacroce, C.; Santilli, F.; Piccoli, L.; Delle Monache, S.; Angelucci, A.; Misasi, R.; Sorice, M.; Mattei, V. Cellular and Molecular Mechanisms Mediated by recPrPC Involved in the Neuronal Differentiation Process of Mesenchymal Stem Cells. Int. J. Mol. Sci. 2019, 20, 345. https://doi.org/10.3390/ijms20020345
Martellucci S, Santacroce C, Santilli F, Piccoli L, Delle Monache S, Angelucci A, Misasi R, Sorice M, Mattei V. Cellular and Molecular Mechanisms Mediated by recPrPC Involved in the Neuronal Differentiation Process of Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2019; 20(2):345. https://doi.org/10.3390/ijms20020345
Chicago/Turabian StyleMartellucci, Stefano, Costantino Santacroce, Francesca Santilli, Luca Piccoli, Simona Delle Monache, Adriano Angelucci, Roberta Misasi, Maurizio Sorice, and Vincenzo Mattei. 2019. "Cellular and Molecular Mechanisms Mediated by recPrPC Involved in the Neuronal Differentiation Process of Mesenchymal Stem Cells" International Journal of Molecular Sciences 20, no. 2: 345. https://doi.org/10.3390/ijms20020345
APA StyleMartellucci, S., Santacroce, C., Santilli, F., Piccoli, L., Delle Monache, S., Angelucci, A., Misasi, R., Sorice, M., & Mattei, V. (2019). Cellular and Molecular Mechanisms Mediated by recPrPC Involved in the Neuronal Differentiation Process of Mesenchymal Stem Cells. International Journal of Molecular Sciences, 20(2), 345. https://doi.org/10.3390/ijms20020345









