Sucrose Synthase Enhances Hull Size and Grain Weight by Regulating Cell Division and Starch Accumulation in Transgenic Rice
Abstract
:1. Introduction
2. Results
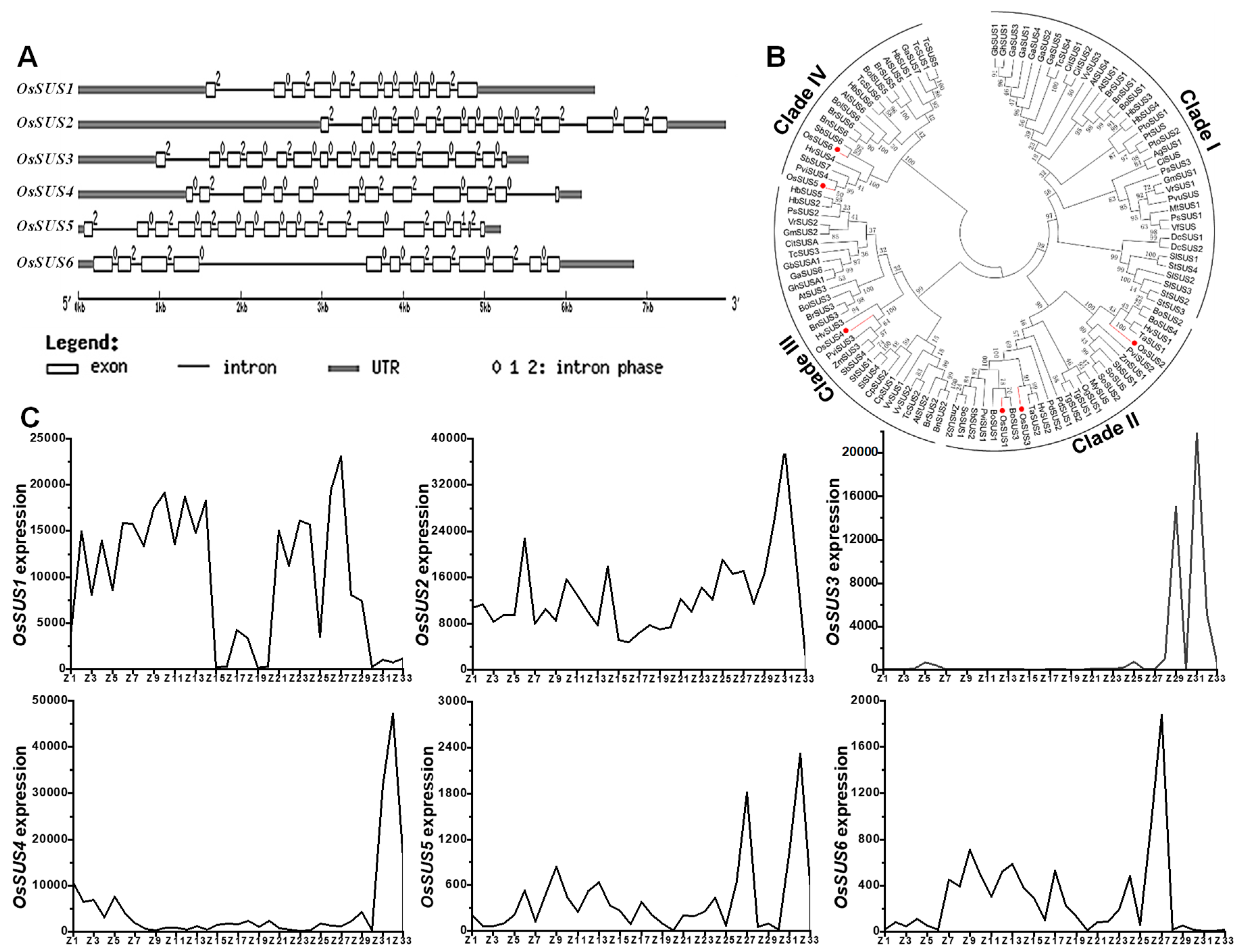
2.1. Phylogenetic and Expression Analyses of OsSUS Family in Rice
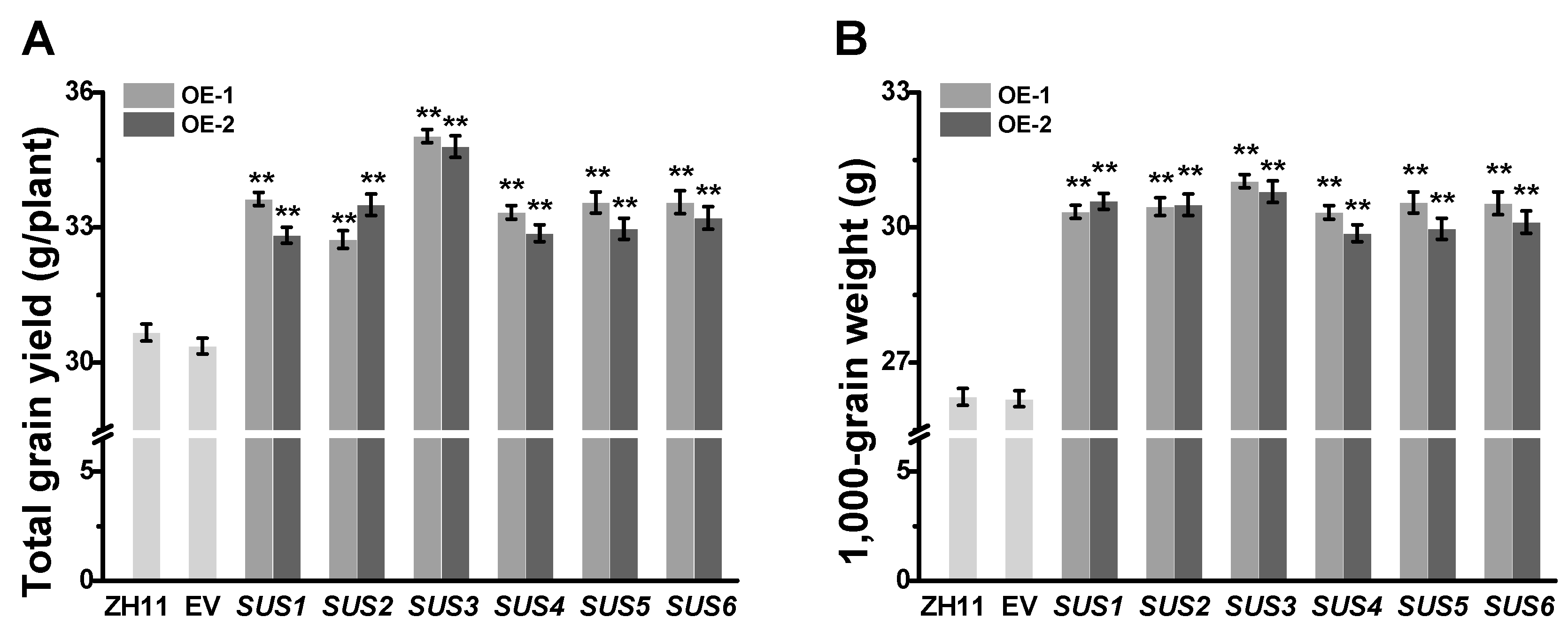
2.2. Increased Grain Weight in All OsSUSs Transgenic Rice Plants
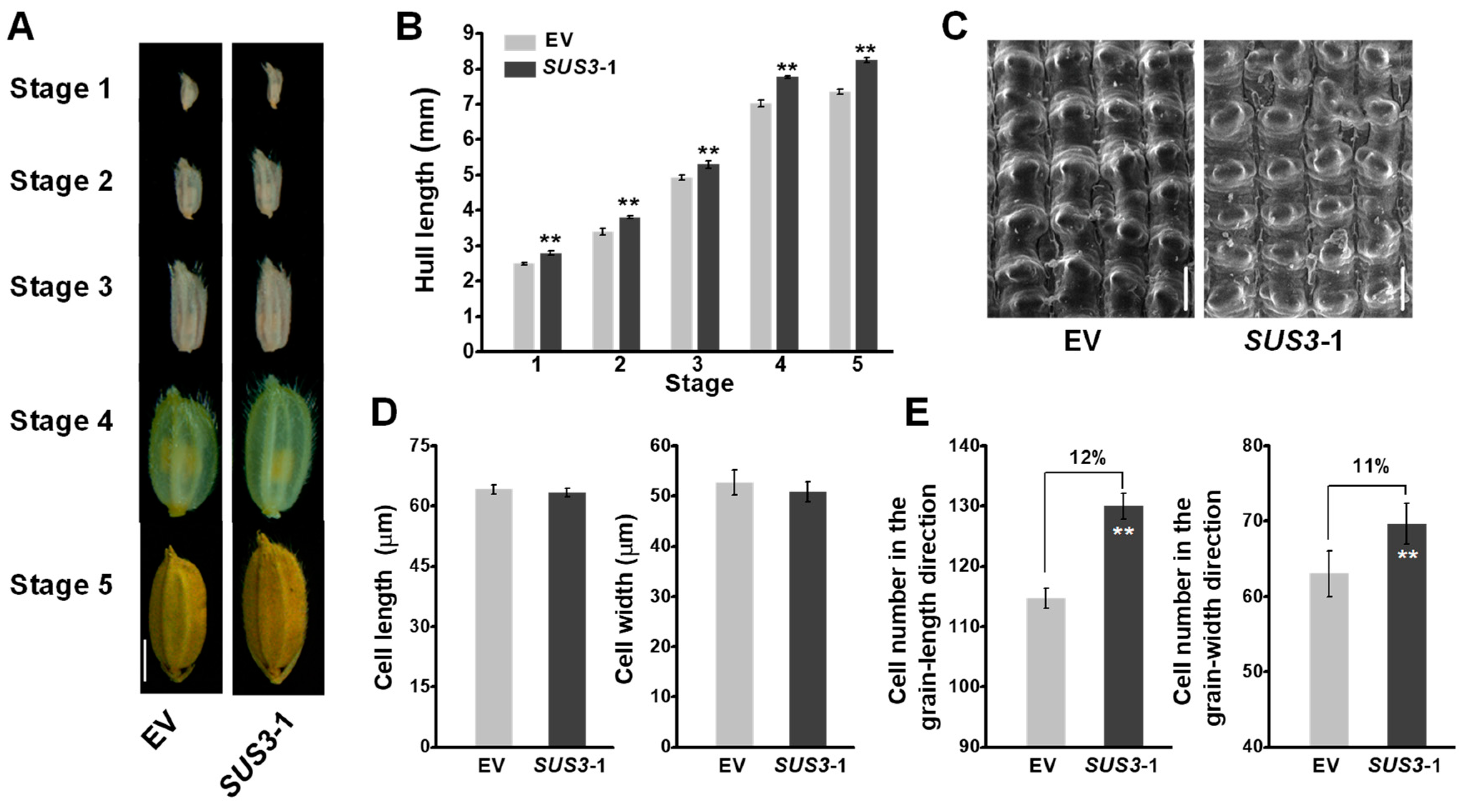
2.3. Raised Grain Length in OsSUS3 Transgenic Lines
2.4. Increased Cell Number in OsSUS3-OE Spikelet Hulls
2.5. Altered Gene Expressions Associated with Cell Division and Cellulose Biosynthesis
2.6. Augmented Starch Accumulation in the OsSUS3 Transgenic Grains
3. Discussion
4. Materials and Methods
4.1. Sequence and Phylogenetic Analysis
4.2. Expression Analysis of OsSUS Gene Family
4.3. Vectors Construction and Gene Transformation
4.4. Field Experiments and Plant Sample Collection
4.5. RNA Preparation and Quantitative PCR
4.6. SUS Enzyme Activity Assay
4.7. Microscope Observation
4.8. Starch Content Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.H.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Li, J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 2014, 48, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [Green Version]
- Qi, P.; Lin, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1; 3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Tong, H.; Shi, B.; Liu, Y.; Fang, S.; Liu, D.; Xiao, Y.; Hu, B.; Liu, L.; Wang, H.; et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2015, 2, 15195. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Li, Y.B.; Fan, C.C.; Xing, Y.Z.; Jiang, Y.H.; Luo, L.J.; Sun, L.; Shao, D.; Xu, C.J.; Li, X.H.; Xiao, J.H.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Duan, P.; Ni, S.; Wang, J.; Zhang, B.; Xu, R.; Wang, Y.; Chen, H.; Zhu, X.; Li, Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2015, 2, 15203. [Google Scholar] [CrossRef]
- Duan, P.; Xu, J.; Zeng, D.; Zhang, B.; Geng, M.; Zhang, G.; Huang, K.; Huang, L.; Xu, R.; Ge, S.; et al. Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 2017, 10, 685–694. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Loboda, T.; Sung, S.J.; Black, C.C. Sucrose synthase in wild tomato, Lycopersicon chmielewskii, and tomato fruit sink strength. Plant Physiol. 1992, 98, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Amor, Y.; Haigler, C.H.; Johnson, S.; Wainscott, M.; Delmer, D.P. A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 1995, 92, 9353–9357. [Google Scholar] [CrossRef] [PubMed]
- Zrenner, R.; Salanoubat, M.; Willmitzer, L.; Sonnewald, U. Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J. 1995, 7, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Sonnewald, U.; Hajirezaei, M.R.; Kossmann, J.; Heyer, A.; Trethewey, R.N.; Willmitzer, L. Increased potato tuber size resulting from apoplastic expression of a yeast invertase. Nat. Biotechnol. 1997, 15, 794–797. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Cheng, W.H.; Taliercio, E.W.; Chourey, P.S. The miniature1 seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 1996, 8, 971–983. [Google Scholar] [CrossRef]
- Liu, J.; Huang, J.; Guo, H.; Lan, L.; Wang, H.; Xu, Y.; Yang, X.; Li, W.; Tong, H.; Xiao, Y.; et al. The conserved and unique genetic architecture of kernel size and weight in maize and rice. Plant Physiol. 2017, 175, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Arioli, T.; Peng, L.; Betzner, A.S.; Burn, J.; Wittke, W.; Herth, W.; Camilleri, C.; Höfte, H.; Plazinski, J.; Birch, R.; et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 1998, 279, 717–720. [Google Scholar] [CrossRef] [PubMed]
- Subbaiah, C.C.; Sachs, M.M. Altered patterns of sucrose synthase phosphorylation and localization precede callose induction and root tip death in anoxic maize seedling. Plant Physiol. 2001, 125, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Baroja-Fernández, E.; Muñoz, F.J.; Montero, M.; Etxeberria, E.; Sesma, M.T.; Ovecka, M.; Bahaji, A.; Ezquer, I.; Li, J.; Prat, S.; et al. Enhancing sucrose synthase activity in transgenic potato (Solanum tuberosum L.) tubers results in increased levels of starch, ADPglucose and UDPglucose and total yield. Plant Cell Physiol. 2009, 50, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, H.; Zhang, Y.; Kang, T.; Zhang, L.; Tong, J.; Xiao, L.; Zhang, H. Constitutive expression of cell wall invertase genes increases grain yield and starch content in maize. Plant Biotechnol. J. 2013, 11, 1080–1091. [Google Scholar] [CrossRef]
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernándeza, E.; Francisco Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E.; et al. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Shiraishi, S.; Tuncel, A.; Matsusaka, H.; Satoh, R.; Singh, S.; Crofts, N.; Hosaka, Y.; Fujita, N.; Hwang, S.; et al. Analysis of the rice ADP-glucose transporter (OsBT1) indicates the presence of regulatory processes in the amyloplast stroma that control ADP-glucose flux into starch. Plant Physiol. 2016, 170, 1271–1283. [Google Scholar] [PubMed]
- Konishi, T.; Ohmiya, Y.; Hayashi, T. Evidence that sucrose loaded into the phloem of a poplar leaf is used directly by sucrose synthase associated with various β-glucan synthases in the stem. Plant Physiol. 2004, 134, 1146–1152. [Google Scholar] [CrossRef]
- Coleman, H.D.; Ellis, D.D.; Gilbert, M.; Mansfield, S.D. Up-regulation of sucrose synthase and UDP-glucose pyrophosphorylase impacts plant growth and metabolism. Plant Biotechnol. J. 2006, 4, 87–101. [Google Scholar] [CrossRef]
- Coleman, H.D.; Yan, J.; Mansfield, S.D. Sucrose synthase affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. USA 2009, 106, 13118–13123. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Guo, W.; Zhu, H.; Ruan, Y.L.; Zhang, T. Overexpression of GhSusA1 increases plant biomass and improves cotton fiber yield and quality. Plant Biotechnol. J. 2012, 10, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.M.; Brill, E.; Llewellyn, D.J.; Furbank, R.T.; Ruan, Y.L. Overexpression of a potato sucrose synthase gene in cotton accelerates leaf expansion, reduces seed abortion, and enhances fiber production. Mol. Plant 2012, 5, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Poovaiah, C.R.; Mazarei, M.; Decker, S.R.; Turner, G.B.; Sykes, R.W.; Davis, M.F.; Stewart, C.N. Transgenic switchgrass (Panicum virgatum L.) biomass is increased by overexpression of switchgrass sucrose synthase (PvSUS1). Biotechnol. J. 2015, 10, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Feng, S.; Huang, J.; Wang, Y.; Wu, L.; Li, X.; Wang, L.; Tu, Y.; Xia, T.; Li, J.; et al. AtCesA8-driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice. Biotechnol. Biofuels 2017, 10, 221. [Google Scholar] [CrossRef] [PubMed]
- Chourey, P.S.; Taliercio, E.W.; Carlson, S.J.; Ruan, Y.L. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 1998, 259, 88–96. [Google Scholar] [PubMed]
- Barratt, D.H.; Derbyshire, P.; Findlay, K.; Pike, M.; Wellner, N.; Lunn, J.; Feil, R.; Simpson, C.; Maule, A.J.; Smith, A.M. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 13124–13129. [Google Scholar] [CrossRef]
- Cho, J.I.; Kim, H.B.; Kim, C.Y.; Hahn, T.R.; Jeon, J.S. Identification and characterization of the duplicate rice sucrose synthase genes OsSUS5 and OsSUS7 which are associated with the plasma membrane. Mol. Cells. 2011, 31, 553–561. [Google Scholar] [CrossRef]
- Cornejo, M.J.; Luth, D.; Blankenship, K.M.; Anderson, O.D.; Blechl, A.E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 1993, 23, 567–581. [Google Scholar] [CrossRef]
- Li, N.; Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016, 33, 23–32. [Google Scholar] [CrossRef]
- Dian, W.; Jiang, H.; Wu, P. Evolution and expression analysis of starch synthase III and IV in rice. J. Exp. Bot. 2005, 56, 623–632. [Google Scholar] [CrossRef] [Green Version]
- Dupuy, L.; Mackenzie, J.; Haseloff, J. Coordination of plant cell division and expansion in a simple morphogenetic system. Proc. Natl. Acad. Sci. USA 2010, 107, 2711–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, D.B.; Cosgrove, D.J. Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 2009, 19, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, C.; Li, Y.; Hu, Z.; Hu, H.; Wang, G.; Li, A.; Wang, Y.; Tu, Y.; Xia, T.; Peng, L.; et al. Ectopic expression of a novel OsExtensin-like gene consistently enhances plant lodging resistance by regulating cell elongation and cell wall thickening in rice. Plant Biotechnol. J. 2018, 16, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Bauer, S.; Brininstool, G.; Facette, M.; Hamann, T.; Milne, J.; Osborne, E.; Paredez, A.; Persson, S.; Raab, T.; et al. Toward a systems approach to understanding plant-cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef]
- Keegstra, K. Plant Cell Walls. Plant Physiol. 2010, 154, 483–486. [Google Scholar] [CrossRef]
- Schuetz, M.; Smith, R.; Ellis, B. Xylem tissue specification, patterning, and differentiation mechanisms. J. Exp. Bot. 2013, 64, 11–31. [Google Scholar] [CrossRef]
- Takehara, K.; Murata, K.; Yamaguchi, T.; Yamaguchi, K.; Chaya, G.; Kido, S.; Iwasaki, Y.; Ogiwara, H.; Ebitani, T.; Miura, K. Thermo-responsive allele of sucrose synthase 3 (Sus3) provides high-temperature tolerance during the ripening stage in rice (Oryza sativa L.). Breed. Sci. 2018, 18007. [Google Scholar] [CrossRef]
- Wang, L.; Xie, W.; Chen, Y.; Tang, W.; Yang, J.; Ye, R.; Liu, L.; Lin, Y.; Xu, C.; Xiao, J.; et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010, 61, 752–766. [Google Scholar] [CrossRef]
- Bieniawska, Z.; Paul Barratt, D.H.; Garlick, A.P.; Thole, V.; Kruger, N.J.; Martin, C.; Zrenner, R.; Smith, A.M. Analysis of the sucrose synthase gene family in Arabidopsis. Plant J. 2007, 49, 810–828. [Google Scholar] [CrossRef] [PubMed]
- Matamoros Fernández, L.E.; Obel, N.; Scheller, H.V.; Roepstorff, P. Differentiation of isomeric oligosaccharide structures by ESI tandem MS and GC-MS. Carbohyd. Res. 2004, 339, 655–664. [Google Scholar] [CrossRef] [PubMed]



| Year | Transgenic Line | Total Grain Yield (g/Plant) | Percentage of Increased Level (%) | 1000-Grain Weight (g) | Percentage of Increased Level (%) | Seeds Per Panicle | Seed Setting Rate (%) | Tillers Per Plant | |
|---|---|---|---|---|---|---|---|---|---|
| Year 1 | ZH11 | ZH11 | 34.01 ± 1.69 | 23.78 ± 0.25 | 133.50 ± 8.94 | 82.18 ± 2.19 | 16.83 ± 1.88 | ||
| Vector | EV | 33.56 ± 2.24 | 23.63 ± 0.48 | 136.33 ± 8.53 | 81.57 ± 1.80 | 16.67 ± 1.03 | |||
| SU3-OE | 1 | 37.56 ± 2.83 * | + 12% † | 25.37 ± 0.37 ** | + 7% | 125.50 ± 5.09 | 82.05 ± 2.43 | 15.67 ± 1.51 | |
| 2 | 36.26 ± 1.47 * | + 8% | 24.77 ± 0.42 ** | + 5% | 123.84 ± 7.99 | 82.21 ± 1.98 | 16.17 ± 1.17 | ||
| 3 | 36.68 ± 2.98 | + 9% | 24.85 ± 0.40 * | + 5% | 124.00 ± 8.83 | 81.49 ± 1.94 | 16.83 ± 0.75 | ||
| 4 | 35.72 ± 1.80 * | + 6% | 25.07 ± 0.27 ** | + 6% | 129.33 ± 6.47 | 83.05 ± 1.53 | 15.50 ± 1.05 | ||
| Year 2 | ZH11 | ZH11 | 27.82 ± 1.14 | 22.92 ± 0.68 | 126.53 ± 4.18 | 80.56 ± 2.94 | 13.50 ± 1.05 | ||
| Vector | EV | 28.44 ± 2.21 | 22.54 ± 0.87 | 129.67 ± 6.38 | 81.50 ± 2.04 | 14.00 ± 0.89 | |||
| SU3-OE | 1 | 33.91 ± 0.77 ** | + 19% | 24.97 ± 1.30 * | + 11% | 123.84 ± 5.04 | 83.24 ± 3.31 | 13.83 ± 0.75 | |
| 2 | 33.90 ± 3.65 ** | + 19% | 23.84 ± 0.53 * | + 6% | 127.33 ± 7.23 | 82.96 ± 3.13 | 13.33 ± 0.52 | ||
| 3 | 31.07 ± 1.27 | + 9% | 24.48 ± 1.08 ** | + 9% | 120.67 ± 6.68 | 81.54 ± 2.22 | 14.00 ± 0.75 | ||
| 4 | 31.39 ± 3.03 * | + 10% | 25.93 ± 0.71 ** | + 15% | 121.17 ± 5.49 | 81.54 ± 1.45 | 13.33 ± 1.03 | ||
| Year 3 | ZH11 | ZH11 | 23.58 ± 0.90 | 23.77 ± 0.04 | 123.83 ± 7.65 | 81.35 ± 2.68 | 10.20 ± 1.15 | ||
| Vector | EV | 23.42 ± 1.25 | 23.76 ± 0.03 | 127.33 ± 7.76 | 81.79 ± 1.70 | 10.50 ± 0.65 | |||
| SU3-OE | 1 | 26.43 ± 0.86 * | +13% | 25.74 ± 0.04 ** | + 8% | 121.00 ± 10.00 | 81.33 ± 2.65 | 11.50 ± 1.01 | |
| 2 | 25.22 ± 0.33 * | +8% | 24.91 ± 0.12 ** | + 5% | 129.67 ± 6.35 | 83.93 ± 0.95 | 11.20 ± 0.82 | ||
| 3 | 25.53 ± 0.87 * | +9% | 25.60 ± 0.10 ** | + 8% | 128.67 ± 5.47 | 83.26 ± 1.96 | 11.40 ± 0.92 | ||
| 4 | 26.68 ± 1.60 ** | +14% | 25.86 ± 0.07 ** | + 9% | 127.50 ± 4.42 | 80.75 ± 3.17 | 10.50 ± 1.05 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, C.; Wang, G.; Wang, Y.; Zhang, R.; Wang, Y.; Feng, S.; Luo, K.; Peng, L. Sucrose Synthase Enhances Hull Size and Grain Weight by Regulating Cell Division and Starch Accumulation in Transgenic Rice. Int. J. Mol. Sci. 2019, 20, 4971. https://doi.org/10.3390/ijms20204971
Fan C, Wang G, Wang Y, Zhang R, Wang Y, Feng S, Luo K, Peng L. Sucrose Synthase Enhances Hull Size and Grain Weight by Regulating Cell Division and Starch Accumulation in Transgenic Rice. International Journal of Molecular Sciences. 2019; 20(20):4971. https://doi.org/10.3390/ijms20204971
Chicago/Turabian StyleFan, Chunfen, Guangya Wang, Youmei Wang, Ran Zhang, Yanting Wang, Shengqiu Feng, Keming Luo, and Liangcai Peng. 2019. "Sucrose Synthase Enhances Hull Size and Grain Weight by Regulating Cell Division and Starch Accumulation in Transgenic Rice" International Journal of Molecular Sciences 20, no. 20: 4971. https://doi.org/10.3390/ijms20204971
APA StyleFan, C., Wang, G., Wang, Y., Zhang, R., Wang, Y., Feng, S., Luo, K., & Peng, L. (2019). Sucrose Synthase Enhances Hull Size and Grain Weight by Regulating Cell Division and Starch Accumulation in Transgenic Rice. International Journal of Molecular Sciences, 20(20), 4971. https://doi.org/10.3390/ijms20204971






