Conjoint Analysis of Genome-Wide lncRNA and mRNA Expression of Heteromorphic Leavesin Response to Environmental Heterogeneityin Populus euphratica
Abstract
1. Introduction
2. Results
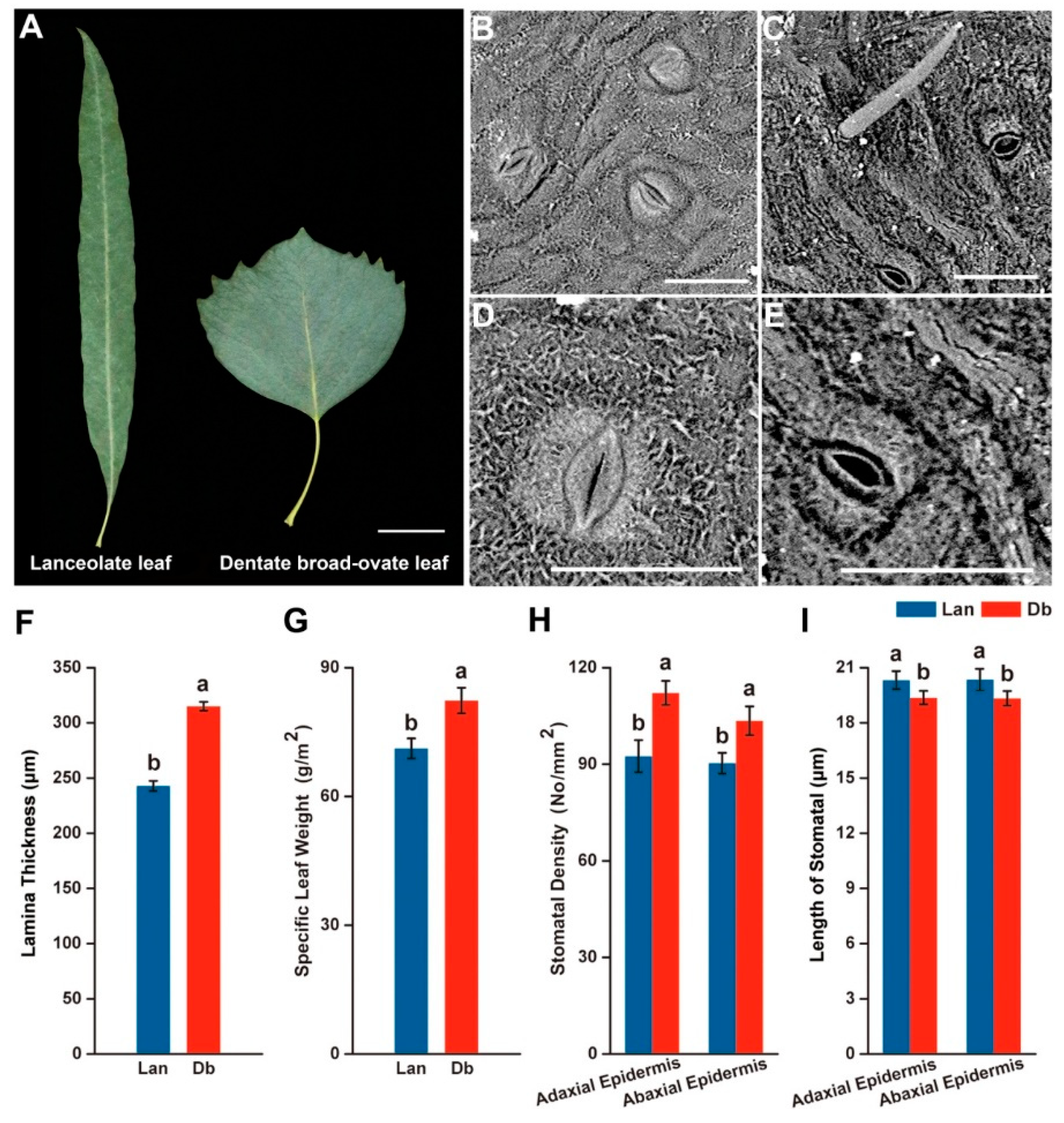
2.1. Morphological Feature of Two Heteromorphic Leaves
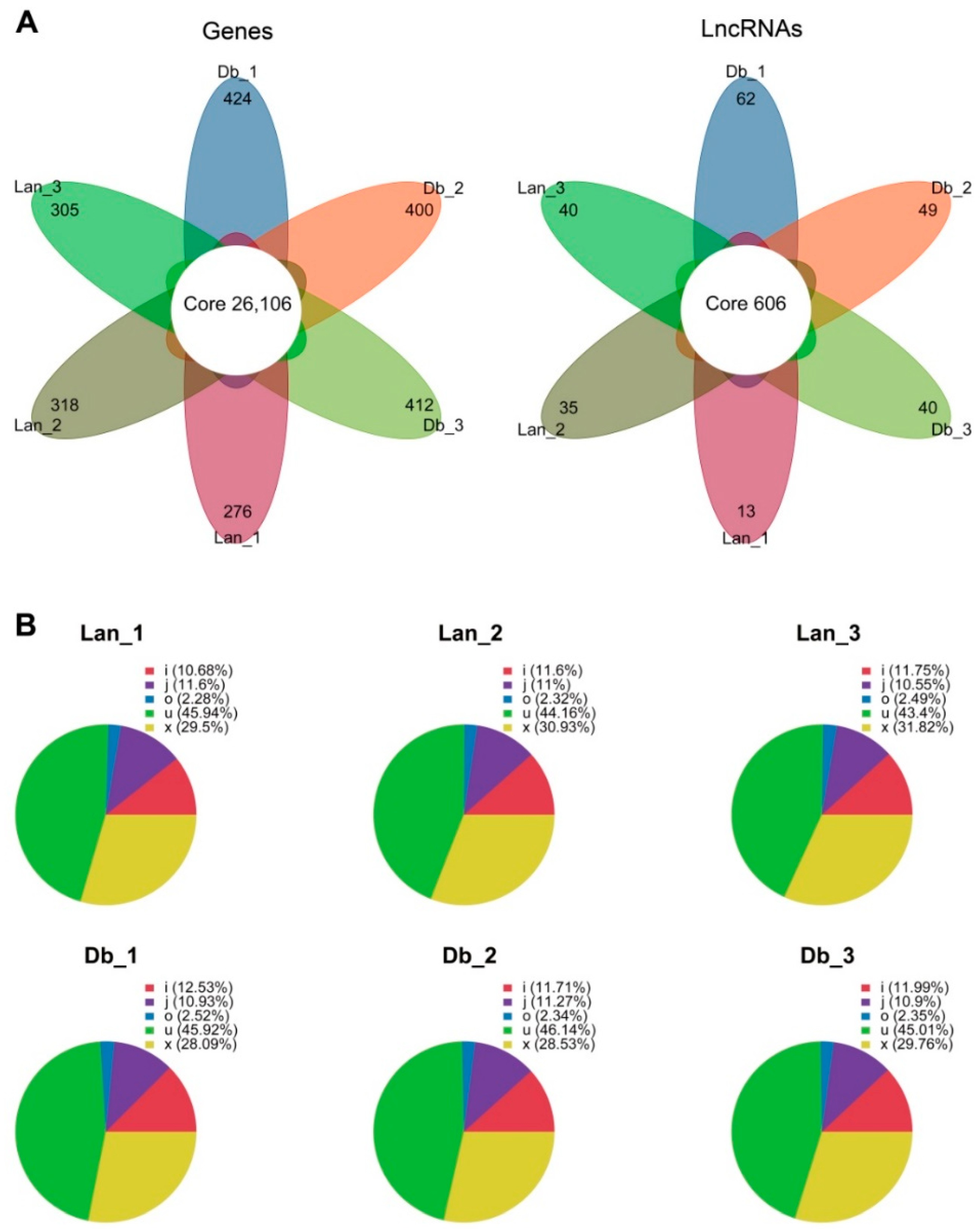
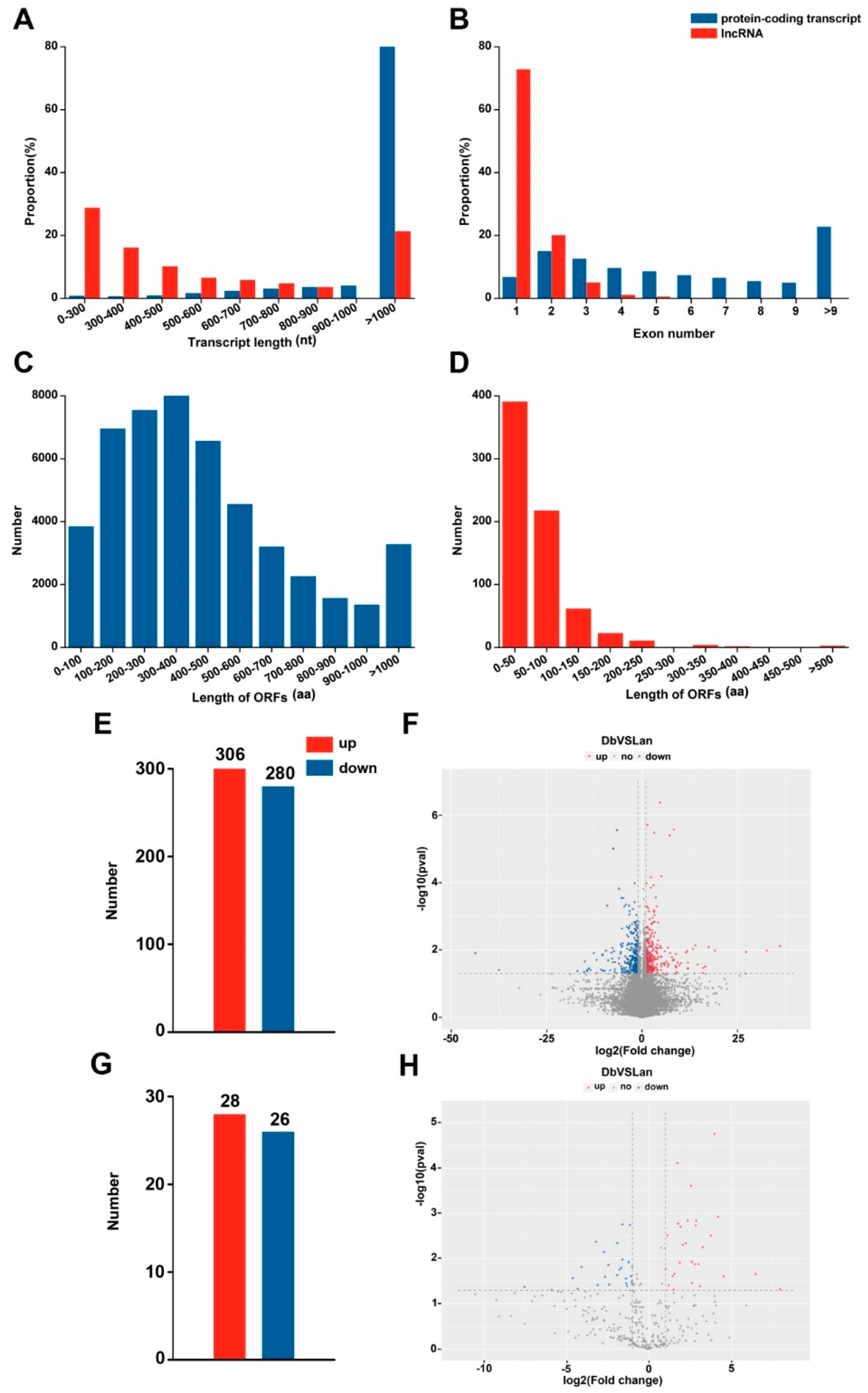
2.2. Global Data Analysis of mRNA and lncRNA Expression in Heteromorphic Leaves
2.3. Identification of Differentially Expressed Genes and lncRNAs
2.4. Identification of Differentially Expressed lncRNA-mRNA Interaction Pairs
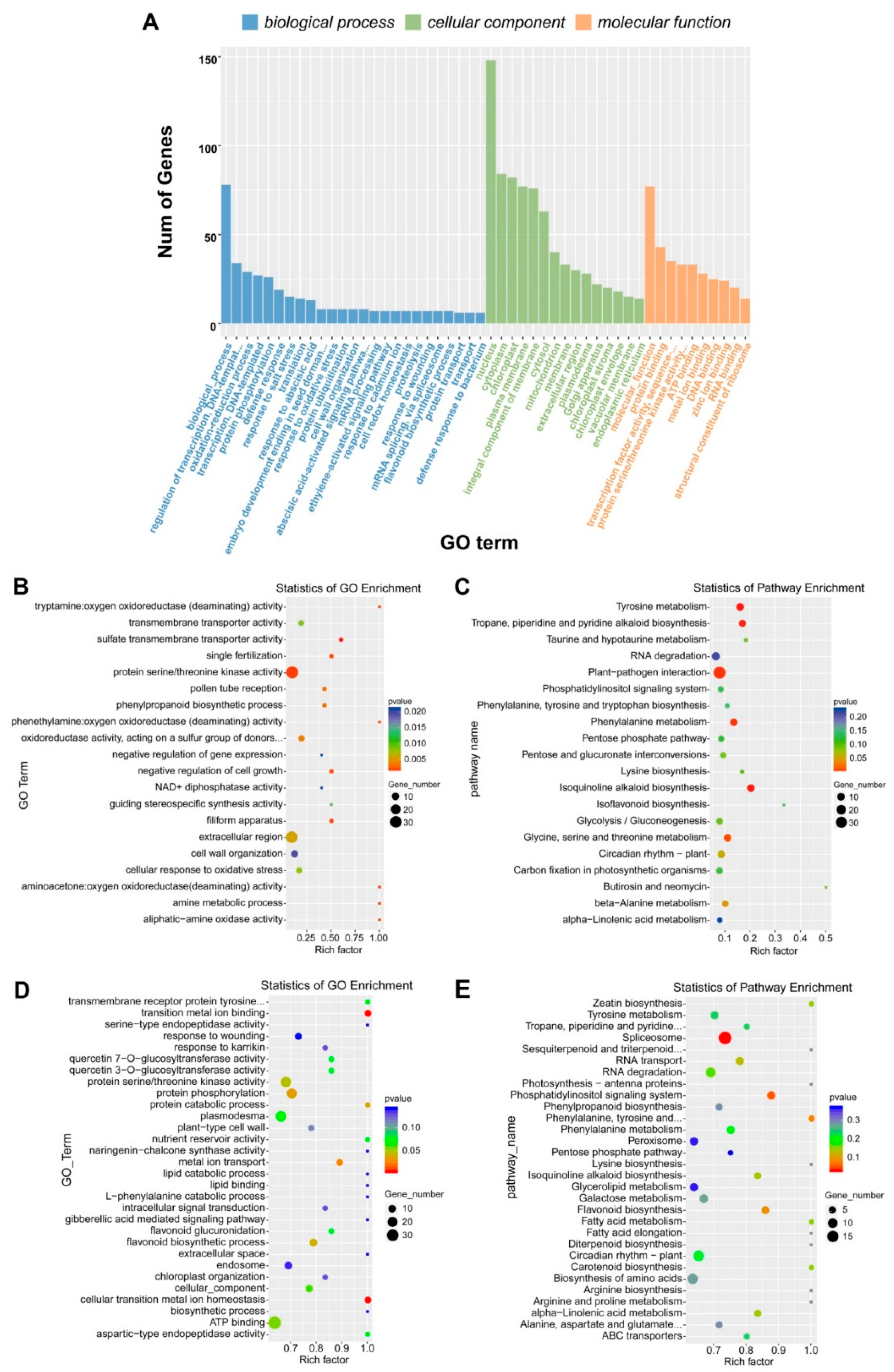
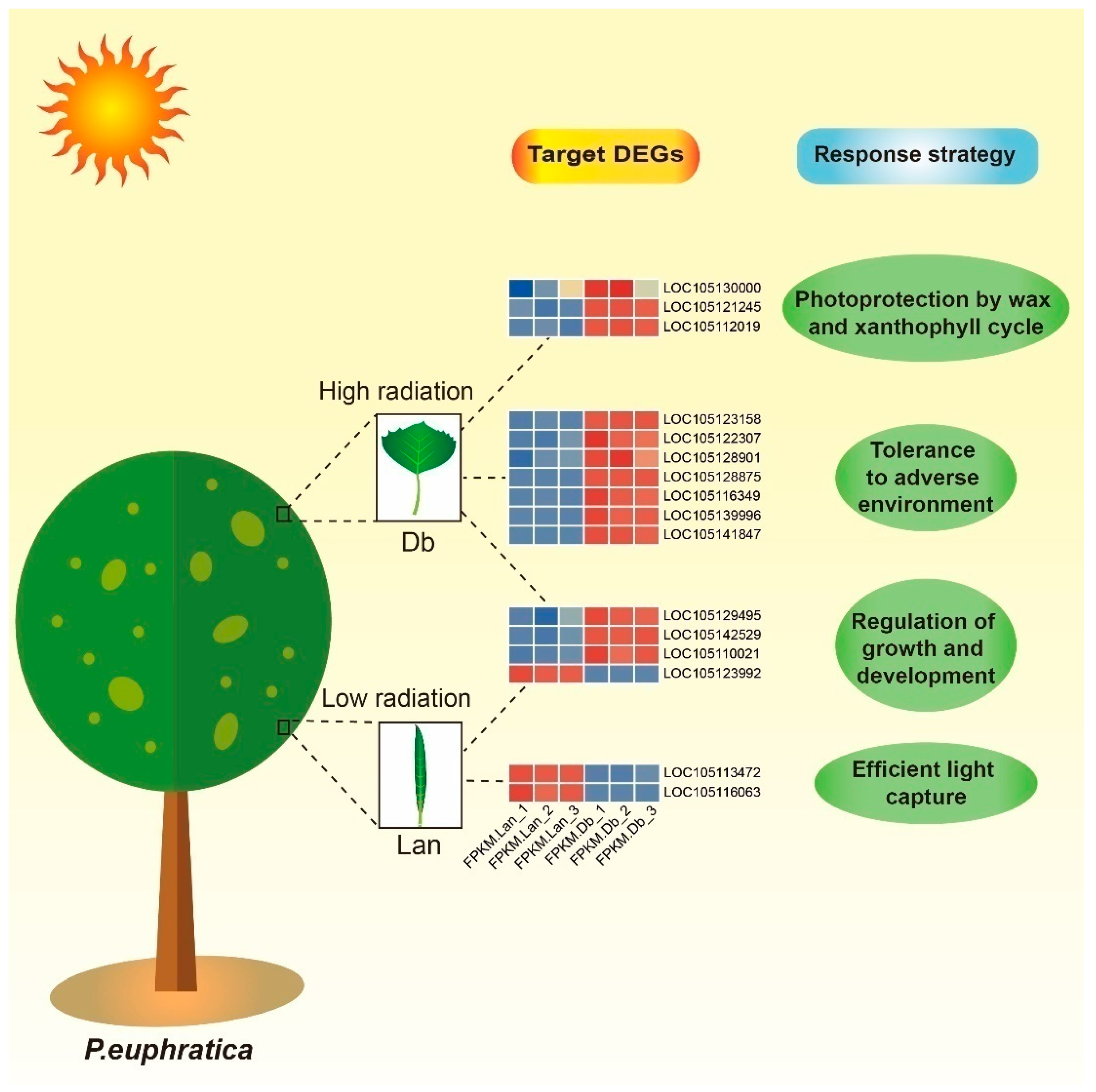
2.5. Interaction of lncRNAs and Target Genes Response to Microenvironment of Heteromorphic Leaves
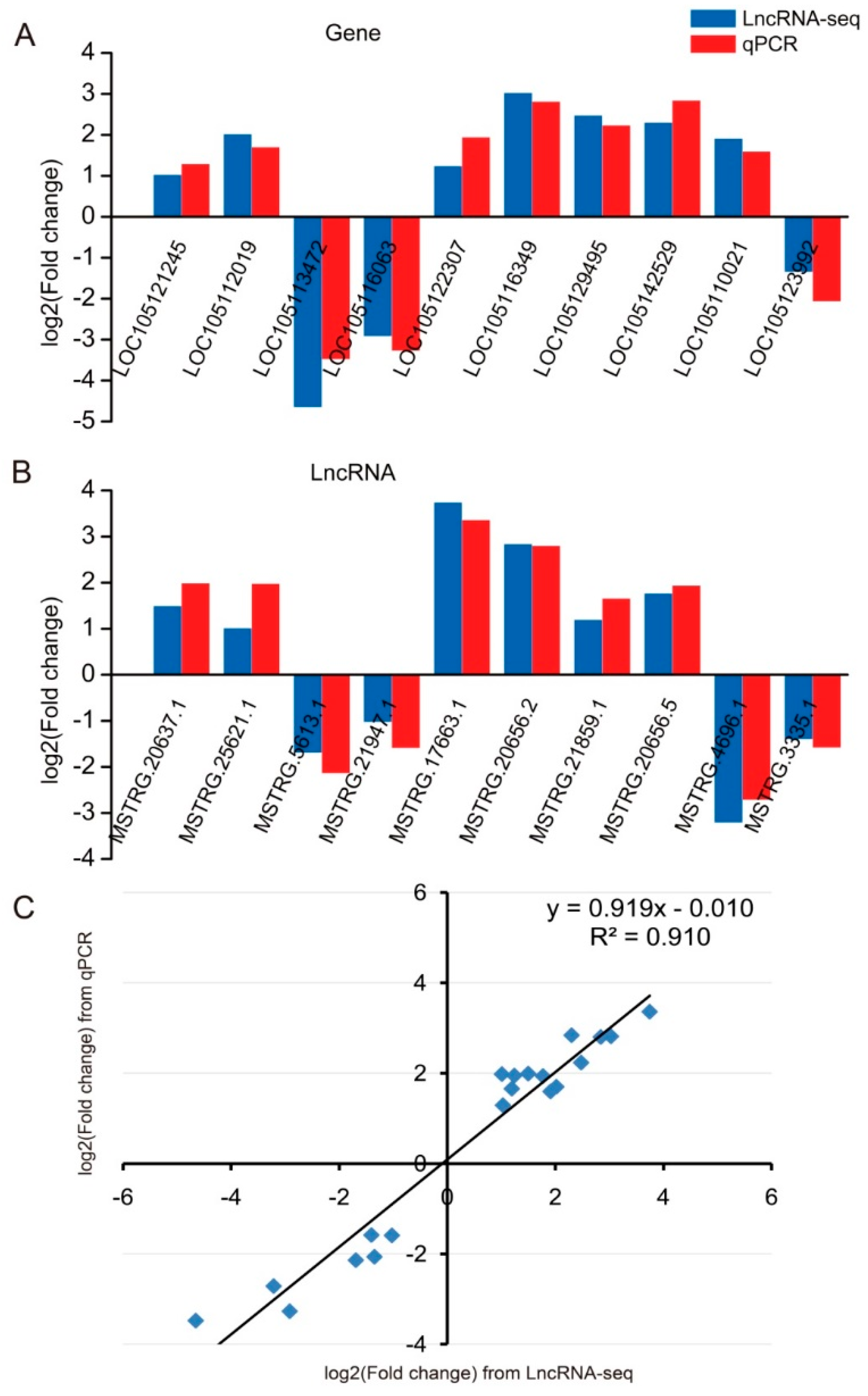
2.6. Validation of qRT-PCR
3. Discussion
3.1. Heteromorphic Leaves Exhibit Different Morphological and Physiological Features to Respond to the Heterogeneous Microenvironments
3.2. Differential Expression Pattern of Genes and lncRNAs in Heteromorphic Leaves
3.3. lncRNA–mRNA Interaction Involved in Response to Light
3.4. lncRNA–mRNA Interaction Involved in Response to Adverse Environment
3.5. lncRNA–mRNA Interaction Involved in Growth and Development
4. Materials and Methods
4.1. Plant Materials
4.2. Measurement of Leaf Morphological and Physiological Features
4.3. cDNA Library Construction and SEQUENCING
4.4. Transcripts Assembly
4.5. lncRNA Identification
4.6. Different Expression Analysis of mRNAs and lncRNAs
4.7. Target Gene Prediction of lncRNAs and Establishment of Coexpression Networks
4.8. Functional Classification of DEGs and the Target DEGs of DELs
4.9. qRT-PCR Validation of lncRNA and Gene Expression Level
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nakayama, H.; Nakayama, N.; Nakamasu, A.; Sinha, N.; Kimura, S. Toward elucidating the mechanisms that regulate heterophylly. Plant Morphol. 2012, 24, 57–63. [Google Scholar] [CrossRef][Green Version]
- Kim, J.; Joo, Y.; Kyung, J.; Jeon, M.; Park, J.Y.; Lee, H.G.; Chung, D.S.; Lee, E.; Lee, I. A molecular basis behind heterophylly in an amphibious plant, Ranunculus trichophyllus. PLoS Genet. 2018, 14, e1007208. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Sinha, N.R.; Kimura, S. How Do Plants and Phytohormones Accomplish Heterophylly, Leaf Phenotypic Plasticity, in Response to Environmental Cues. Front. Plant Sci. 2017, 8, 1777. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Guo, P.; Gugger, P.F.; Guo, Y.; Liu, X.; Chen, J. Investigating the molecular basis for heterophylly in the aquatic plant Potamogeton octandrus (Potamogetonaceae) with comparative transcriptomics. PeerJ 2018, 6, e4448. [Google Scholar] [CrossRef] [PubMed]
- Zotz, G.; Wilhelm, K.; Becker, A. Heteroblasty—A Review. Bot. Rev. 2011, 77, 109–151. [Google Scholar] [CrossRef]
- Kordyum, E.; Klimenko, E. Chloroplast ultrastructure and chlorophyll performance in the leaves of heterophyllous Nuphar lutea (L.) Smith. plants. Aquat. Bot. 2013, 110, 84–91. [Google Scholar] [CrossRef]
- Nakayama, H.; Kimura, S. Leaves may function as temperature sensors in the heterophylly of Rorippa aquatica (Brassicaceae). Plant Signal. Behav. 2015, 10, e1091909. [Google Scholar] [CrossRef]
- Li, G.; Hu, S.; Yang, J.; Schultz, E.A.; Clarke, K.; Hou, H. Water-Wisteria as an ideal plant to study heterophylly in higher aquatic plants. Plant Cell Rep. 2017, 36, 1225–1236. [Google Scholar] [CrossRef]
- Hao, J.; Yue, N.; Zheng, C. Analysis of changes in anatomical characteristics and physiologic features of heteromorphic leaves in a desert tree, Populus euphratica. Acta Physiol. Plant 2017, 39, 160. [Google Scholar] [CrossRef]
- Leigh, A.; Zwieniecki, M.A.; Rockwell, F.E.; Boyce, C.K.; Nicotra, A.B.; Holbrook, N.M. Structural and hydraulic correlates of heterophylly in Ginkgo biloba. New Phytol. 2011, 189, 459–470. [Google Scholar] [CrossRef]
- Xiao, H.; Wang, C.; Liu, J.; Wang, L.; Du, D. Insights into the differences in leaf functional traits of heterophyllous Syringa oblata under different light intensities. J. For. Res. 2015, 26, 613–621. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, Evolution, and Mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yuan, D.; Tu, L.; Gao, W.; He, Y.; Hu, H.; Wang, P.; Liu, N.; Lindsey, K.; Zhang, X. Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytol. 2015, 207, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.; Wu, Y.; Li, J.; Lin, H.; Hu, L.; Zhang, T.; Qi, Y.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Zhu, Q.H.; Stephen, S.; Taylor, J.; Helliwell, C.A.; Wang, M.B. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytol. 2014, 201, 574–584. [Google Scholar] [CrossRef]
- Xu, X.W.; Zhou, X.H.; Wang, R.R.; Peng, W.L.; An, Y.; Chen, L.L. Functional analysis of long intergenic non-coding RNAs in phosphate-starved rice using competing endogenous RNA network. Sci. Rep. 2016, 6, 20715. [Google Scholar] [CrossRef]
- Fan, C.; Hao, Z.; Yan, J.; Li, G. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genom. 2015, 16, 793. [Google Scholar] [CrossRef]
- Chen, M.; Wang, C.; Bao, H.; Chen, H.; Wang, Y. Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genom. 2016, 291, 1663–1680. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zhang, X.; Wang, W.; Yuan, R.; Shen, F. Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol. 2018, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Li, J.; Yang, Y.; Tan, C.; Zhu, Y.; Hu, L.; Qi, Y.; Lu, Z.J. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 2018, 93, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Luan, Y.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef]
- Wang, R.; Zou, J.; Meng, J.; Wang, J. Integrative analysis of genome-wide lncRNA and mRNA expression in newly synthesized Brassica hexaploids. Ecol. Evolut. 2018, 8, 6034–6052. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Chen, G.; Li, M.; Liu, M.; Liu, D. Epidermal Micromorphology and Mesophyll Structure of Populus euphratica Heteromorphic Leaves at Different Development Stages. PLoS ONE 2015, 10, e0137701. [Google Scholar] [CrossRef]
- Zheng, C.; Qiu, J.; Jiang, C.; Yue, N.; Wang, X.; Wang, W. Comparison of stomatal characteristics and photosynthesis of polymorphic Populus euphratica leaves. Front. For. China 2007, 2, 87–93. [Google Scholar] [CrossRef]
- Liu, X.; Chang, Z.; Ma, Y.; Wu, Y. Characteristics of the Fast Chlorophyll Fluorescence Induction Kinetics of Heteromorphic Leaves in Populus euphratica. J. Desert Res. 2014, 34, 704–711. [Google Scholar]
- Chen, Z.Y.; Peng, Z.S.; Yang, J.; Chen, W.Y.; Ou-Yang, Z.M. A mathematical model for describing light-response curves in Nicotiana tabacum L. Photosynthetica 2011, 49, 467. [Google Scholar] [CrossRef]
- Craine, J.M.; Reich, P.B. Leaf-level light compensation points in shade-tolerant woody seedlings. New Phytol. 2005, 166, 710–713. [Google Scholar] [CrossRef]
- Zhong, S.; Shi, H.; Xue, C.; Wang, L.; Xi, Y.; Li, J.; Quail, P.H.; Deng, X.W.; Guo, H. A Molecular Framework of Light-Controlled Phytohormone Action in Arabidopsis. Curr. Biol. 2012, 22, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Valiente, J.A.; Sánchez-Gómez, D.; Aranda, I.; Valladares, F. Phenotypic plasticity and local adaptation in leaf ecophysiological traits of 13 contrasting cork oak populations under different water availabilities. Tree Physiol. 2010, 30, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Mojzes, A.; Kalapos, T.; Virágh, K. Plasticity of leaf and shoot morphology and leaf photochemistry for Brachypodium pinnatum (L.) Beauv. growing in contrasting microenvironments in a semiarid loess forest-steppe vegetation mosaic. Flora Morphol. Distrib. Funct. Ecol. Plants 2003, 198, 304–320. [Google Scholar] [CrossRef]
- Tattini, M.; Gravano, E.; Pinelli, P.; Mulinacci, N.; Romani, A. Flavonoids accumulate in leaves and glandular trichomes of Phillyrea latifolia exposed to excess solar radiation. New Phytol. 2000, 148, 69–77. [Google Scholar] [CrossRef]
- Guo, P.; Wei, H.; Zhang, W.; Bao, Y. Physiological Responses of Alfalfa to High-level Salt Stress: Root Ion Flux and Stomatal Characteristics. Int. J. Agric. Biol. 2015, 18, 125–133. [Google Scholar] [CrossRef]
- Cohu, C.M.; Muller, O.; Adams, W.W., III; Demmig-Adams, B. Leaf anatomical and photosynthetic acclimation to cool temperature and high light in two winter versus two summer annuals. Physiol. Plant 2014, 152, 164–173. [Google Scholar] [CrossRef]
- Puglielli, G.; Varone, L.; Gratani, L.; Catoni, R. Specific leaf area variations drive acclimation of Cistus salvifolius in different light environments. Photosynthetica 2017, 55, 31–40. [Google Scholar] [CrossRef]
- Urban, O.; Košvancová, M.; Marek, M.V.; Lichtenthaler, H.K. Induction of photosynthesis and importance of limitations during the induction phase in sun and shade leaves of five ecologically contrasting tree species from the temperate zone. Tree Physiol. 2007, 27, 1207–1215. [Google Scholar] [CrossRef]
- BABA-KASAI, A.; Hara, N.; Takano, M. Tissue-specific and light-dependent regulation of phytochrome gene expression in rice. Plant Cell Environ. 2014, 37, 2654–2666. [Google Scholar] [CrossRef]
- Yu, L.; Ma, J.; Niu, Z.; Bai, X.; Lei, W.; Shao, X.; Chen, N.; Zhou, F.; Wan, D. Tissue-Specific Transcriptome Analysis Reveals Multiple Responses to Salt Stress in Populus euphratica Seedlings. Genes 2017, 8, 372. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2013, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wu, F.; Yan, Z.; Li, J.; Ma, T.; Zhang, Y.; Zhao, Y.; Wang, Y.; Zhang, J. Differential co-expression networks of long non-coding RNAs and mRNAs in Cleistogenes songorica under water stress and during recovery. BMC Plant Biol. 2019, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xia, X.; Jiang, H.; Lu, Z.; Cui, J.; Cao, F.; Jin, B. Genome-wide identification and characterization of novel lncRNAs in Ginkgo biloba. Trees 2018, 32, 1429–1442. [Google Scholar] [CrossRef]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.; Zhang, W.H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Van Wittenberghe, S.; Adriaenssens, S.; Staelens, J.; Verheyen, K.; Samson, R. Variability of stomatal conductance, leaf anatomy, and seasonal leaf wettability of young and adult European beech leaves along a vertical canopy gradient. Trees 2012, 26, 1427–1438. [Google Scholar] [CrossRef]
- Anderson, J.M.; Park, Y.I.; Chow, W.S. Photoinactivation and photoprotection of photosystem II in nature. Physiol. Plant 1997, 100, 214–223. [Google Scholar] [CrossRef]
- Ort, D.R. When There Is Too Much Light. Plant Physiol. 2001, 125, 29–32. [Google Scholar] [CrossRef]
- Shepherd, T.; Wynne Griffiths, D. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef]
- Holmes, M.G.; Keiller, D.R. Effects of pubescence and waxes on the reflectance of leaves in the ultraviolet and photosynthetic wavebands: A comparison of a range of species. Plant Cell Environ. 2002, 25, 85–93. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing Plant Surfaces: Cuticular Wax Formation by Epidermal Cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef]
- Todd, J.; Post-Beittenmiller, D.; Jaworski, J.G. KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 1999, 17, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ranathunge, K.; Huang, H.; Pei, Z.; Franke, R.; Schreiber, L.; He, C. Wax Crystal-Sparse Leaf1 encodes a β–ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 2008, 228, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Laila, R.; Robin, A.H.K.; Yang, K.; Park, J.I.; Suh, M.C.; Kim, J.; Nou, I.S. Developmental and Genotypic Variation in Leaf Wax Content and Composition, and in Expression of Wax Biosynthetic Genes in Brassica oleracea var. capitata. Front. Plant Sci. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.A.; Lovelock, C.E.; Osmond, C.B. Wax as a Mechanism for Protection against Photoinhibition—A Study of Cotyledon orbiculata. Bot. Acta 1993, 106, 307–312. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 2009, 12, 721–727. [Google Scholar] [CrossRef]
- Gilmore, A.M. Mechanistic aspects of xanthophyll cycle-dependent photoprotection in higher plant chloroplasts and leaves. Physiol. Plant 1997, 99, 197–209. [Google Scholar] [CrossRef]
- Song, X.S.; Shang, Z.W.; Yin, Z.P.; Ren, J.; Sun, M.C.; Ma, X.L. Mechanism of xanthophyll-cycle-mediated photoprotection in Cerasus humilis seedlings under water stress and subsequent recovery. Photosynthetica 2011, 49, 523–530. [Google Scholar] [CrossRef]
- Umate, P. Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in Arabidopsis and rice. Plant Signal. Behav. 2010, 5, 1537–1542. [Google Scholar] [CrossRef]
- Szymańska, R.; Ślesak, I.; Orzechowska, A.; Kruk, J. Physiological and biochemical responses to high light and temperature stress in plants. Environ. Exp. Bot. 2017, 139, 165–177. [Google Scholar] [CrossRef]
- Nawkar, G.M.; Maibam, P.; Park, J.H.; Sahi, V.P.; Lee, S.Y.; Kang, C.H. UV-Induced Cell Death in Plants. Int. J. Mol. Sci. 2013, 14, 1608–1628. [Google Scholar] [CrossRef]
- Hideg, É.; Jansen, M.A.K.; Strid, Å. UV-B exposure, ROS, and stress: Inseparable companions or loosely linked associates? Trends Plant Sci. 2013, 18, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Korovila, I.; Hugo, M.; Castro, J.P.; Weber, D.; Höhn, A.; Grune, T.; Jung, T. Proteostasis, oxidative stress and aging. Redox Biol. 2017, 13, 550–567. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, T.; Heinemann, S. Regulation of cell function by methionine oxidation and reduction. J. Physiol. 2010, 531, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kurek, I.; Erel, N.; Herman, E.A.; Aviezer, K. The wheat peptidyl prolyl cis-trans-isomerase FKBP77 is heat induced and developmentally regulated. Plant Physiol. 1999, 119, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.D.; Wajapeyee, N.; Yadav, V.; Singh, P. Stress-Induced Changes in Peptidyl-Prolyl cis-trans Isomerase Activity of Sorghum bicolor Seedlings. Biol. Plant 2003, 47, 367–371. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Et Biophys. Acta Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Li, S.; Zhou, X.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 2010, 29, 475–483. [Google Scholar] [CrossRef]
- Muthusamy, M.; Uma, S.; Backiyarani, S.; Saraswathi, M.S. Genome-wide screening for novel, drought stress-responsive long non-coding RNAs in drought-stressed leaf transcriptome of drought-tolerant and -susceptible banana (Musa spp) cultivars using Illumina high-throughput sequencing. Plant Biotechnol. Rep. 2015, 9, 279–286. [Google Scholar] [CrossRef]
- Pitorre, D.; Llauro, C.; Jobet, E.; Guilleminot, J.; Brizard, J.P.; Delseny, M.; Lasserre, E. RLK7, a leucine-rich repeat receptor-like kinase, is required for proper germination speed and tolerance to oxidative stress in Arabidopsis thaliana. Planta 2010, 232, 1339–1353. [Google Scholar] [CrossRef]
- Zhou, F.; Guo, Y.; Qiu, L.J. Genome-wide identification and evolutionary analysis of leucine-rich repeat receptor-like protein kinase genes in soybean. BMC Plant Biol. 2016, 16, 58. [Google Scholar] [CrossRef]
- Iqbal, A.; Wang, T.; Wu, G.; Tang, W.; Zhu, C.; Wang, D.; Li, Y.; Wang, H. Physiological and transcriptome analysis of heteromorphic leaves and hydrophilic roots in response to soil drying in desert Populus euphratica. Sci. Rep. 2017, 7, 12188. [Google Scholar] [CrossRef] [PubMed]
- Whitelam, G.C.; Devlin, P.F. Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant. Cell Environ. 2008, 20, 752–758. [Google Scholar] [CrossRef]
- Park, D.H.; Somers, D.E.; Kim, Y.S.; Choy, Y.H.; Lim, H.K.; Soh, M.S.; Kim, H.J.; Kay, S.A.; Nam, H.G. Control of Circadian Rhythms and Photoperiodic Flowering by the Arabidopsis GIGANTEA Gene. Science 1999, 285, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Kawahigashi, H.; Oshima, M.; Ando, T.; Handa, H. The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J. Exp. Bot. 2011, 63, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Hettiarachchi, G.H.C.M.; Deng, X.W.; Holm, M. Arabidopsis CONSTANS-LIKE3 Is a Positive Regulator of Red Light Signaling and Root Growth. Plant Cell 2006, 18, 70–84. [Google Scholar] [CrossRef] [PubMed]
- Yokawa, K.; Koshiba, T.; Baluška, F. Light-dependent control of redox balance and auxin biosynthesis in plants. Plant Signal. Behav. 2014, 9, e29522. [Google Scholar] [CrossRef] [PubMed]
- Kardel, F.; Wuyts, K.; Babanezhad, M.; Vitharana, U.W.A.; Wuytack, T.; Potters, G.; Samson, R. Assessing urban habitat quality based on specific leaf area and stomatal characteristics of Plantago lanceolata L. Environ. Pollut. 2010, 158, 788–794. [Google Scholar] [CrossRef]
- Atici, Ö.; Ağar, G.; Battal, P. Changes in phytohormone contents in chickpea seeds germinating under lead or zinc stress. Biol. Plant 2005, 49, 215–222. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511. [Google Scholar] [CrossRef]
- Liao, Q.; Liu, C.; Yuan, X.; Kang, S.; Miao, R.; Xiao, H.; Zhao, G.; Luo, H.; Bu, D.; Zhao, H.; et al. Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Res. 2011, 39, 3864–3878. [Google Scholar] [CrossRef]
- Chen, J.; Quan, M.; Zhang, D. Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 2015, 241, 125–143. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069. [Google Scholar] [CrossRef] [PubMed]
- Huntley, R.; Dimmer, E.; Barrell, D.; Binns, D.; Apweiler, R. The Gene Ontology Annotation (GOA) Database. Nat. Preced. 2009. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Li, L.; Tang, S.; Yuan, C.; Tian, Q.; Su, Y.; Li, H.G.; Zhao, L.; Yin, W.; Zhao, R.; et al. Evaluation of Appropriate Reference Genes for Reverse Transcription-Quantitative PCR Studies in Different Tissues of a Desert Poplar via Comparision of Different Algorithms. Int. J. Mol. Sci. 2015, 16, 20468–20491. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Sample | Raw Data | Valid Data | Q20% | Q30% | GC Content% | ||
|---|---|---|---|---|---|---|---|
| Read | Base | Read | Base | ||||
| Lan_1 | 191446580 | 28.72G | 135085944 | 20.26G | 99.70 | 95.40 | 43.50 |
| Lan_2 | 166688106 | 25.00G | 112481178 | 16.87G | 99.75 | 95.83 | 43.50 |
| Lan_3 | 165831618 | 24.87G | 110553910 | 16.58G | 99.64 | 95.00 | 43.50 |
| Db_1 | 163034286 | 24.46G | 99461962 | 14.92G | 99.68 | 95.73 | 44 |
| Db_2 | 165850472 | 24.88G | 111601846 | 16.74G | 99.66 | 94.90 | 44 |
| Db_3 | 172292388 | 25.84G | 118538356 | 17.78G | 99.65 | 95.60 | 44 |
| Sample | Lan_1 | Lan_2 | Lan_3 | Db_1 | Db_2 | Db_3 | Total |
|---|---|---|---|---|---|---|---|
| Expressed Gene | 31,392 | 31,293 | 31,410 | 31,399 | 31,307 | 31,543 | 36,492 |
| Expressed lncRNA | 1095 | 1164 | 1166 | 1189 | 1153 | 1193 | 1725 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, M.; He, S.; Hao, L.; Li, Y.; Zheng, C.; Zhao, Y. Conjoint Analysis of Genome-Wide lncRNA and mRNA Expression of Heteromorphic Leavesin Response to Environmental Heterogeneityin Populus euphratica. Int. J. Mol. Sci. 2019, 20, 5148. https://doi.org/10.3390/ijms20205148
Zeng M, He S, Hao L, Li Y, Zheng C, Zhao Y. Conjoint Analysis of Genome-Wide lncRNA and mRNA Expression of Heteromorphic Leavesin Response to Environmental Heterogeneityin Populus euphratica. International Journal of Molecular Sciences. 2019; 20(20):5148. https://doi.org/10.3390/ijms20205148
Chicago/Turabian StyleZeng, Ming, Shuhang He, Lin Hao, Yujie Li, Caixia Zheng, and Yuanyuan Zhao. 2019. "Conjoint Analysis of Genome-Wide lncRNA and mRNA Expression of Heteromorphic Leavesin Response to Environmental Heterogeneityin Populus euphratica" International Journal of Molecular Sciences 20, no. 20: 5148. https://doi.org/10.3390/ijms20205148
APA StyleZeng, M., He, S., Hao, L., Li, Y., Zheng, C., & Zhao, Y. (2019). Conjoint Analysis of Genome-Wide lncRNA and mRNA Expression of Heteromorphic Leavesin Response to Environmental Heterogeneityin Populus euphratica. International Journal of Molecular Sciences, 20(20), 5148. https://doi.org/10.3390/ijms20205148




