A Genome-Centric Approach Reveals a Novel Glycosyltransferase from the GA A07 Strain of Bacillus thuringiensis Responsible for Catalyzing 15-O-Glycosylation of Ganoderic Acid A
Abstract
1. Introduction
2. Results
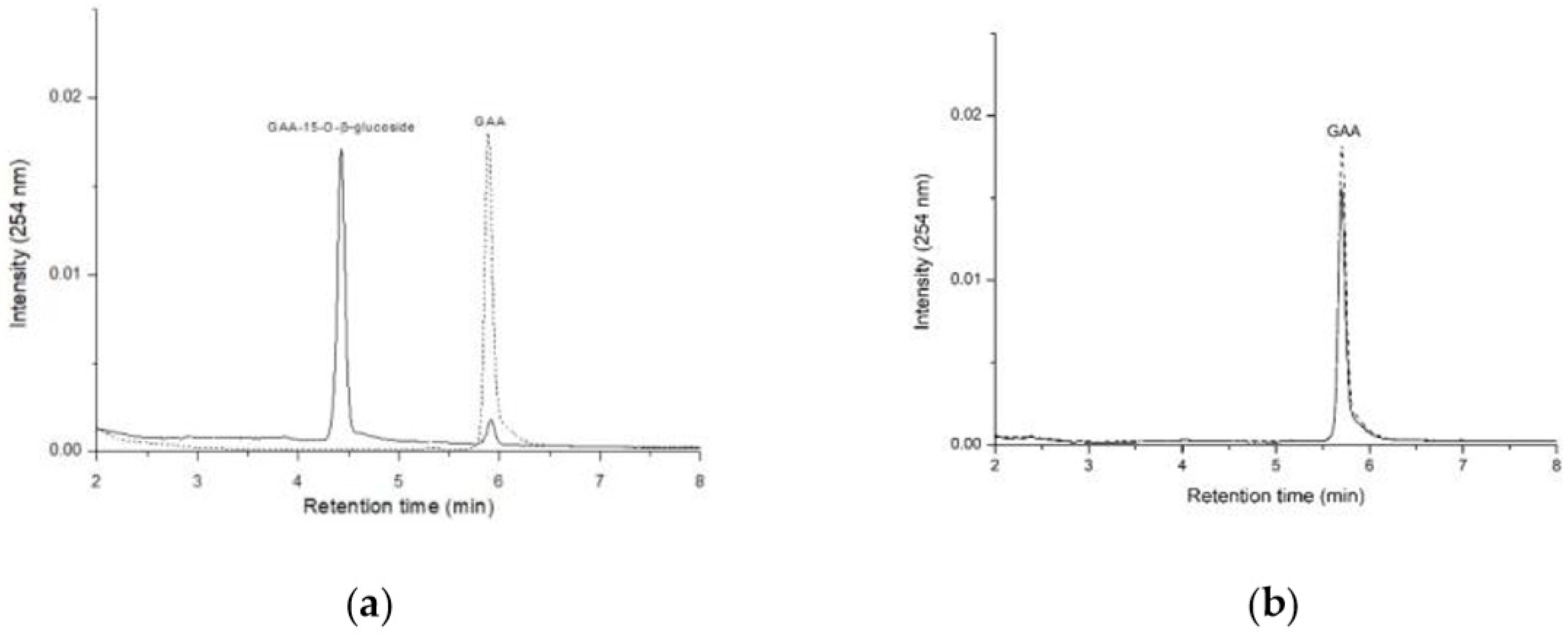
2.1. Comparison of GAA-15-O-β-Glucoside Production between B. subtilis ATCC 6633 and Bacillus sp. GA A07
2.2. Genome Sequencing, Assembly, Annotation, and Reclassification of the GA A07 Strain
2.3. Phylogenetic Analysis of GTs from the GA A07 Strain
2.4. Cloning, Overexpression, and Purification of GT from the GA A07 Strain in E. coli
2.5. Activity Assays of Recombinant GT Proteins toward GAA
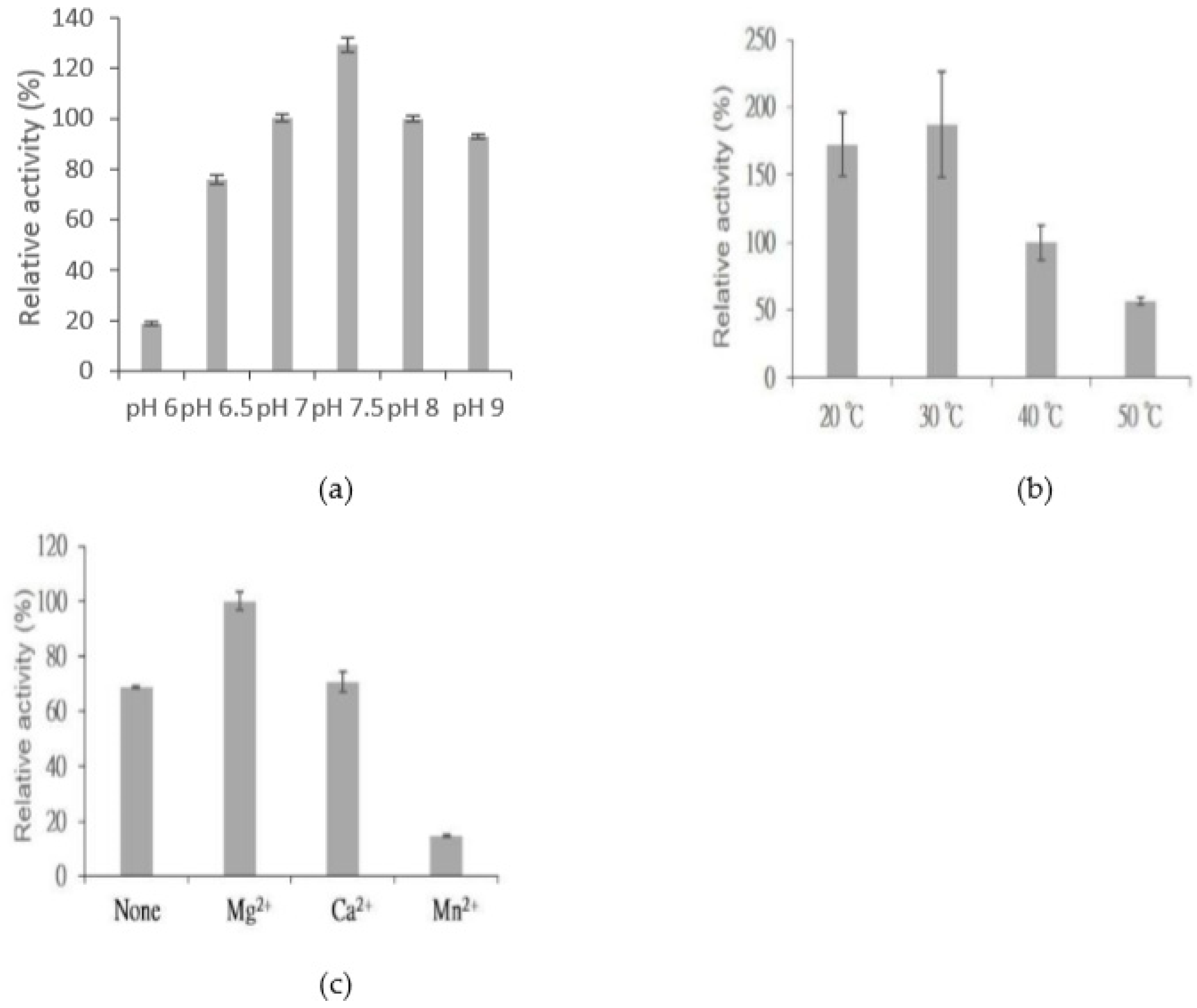
2.6. Catalytic Conditions for BtGT_16345
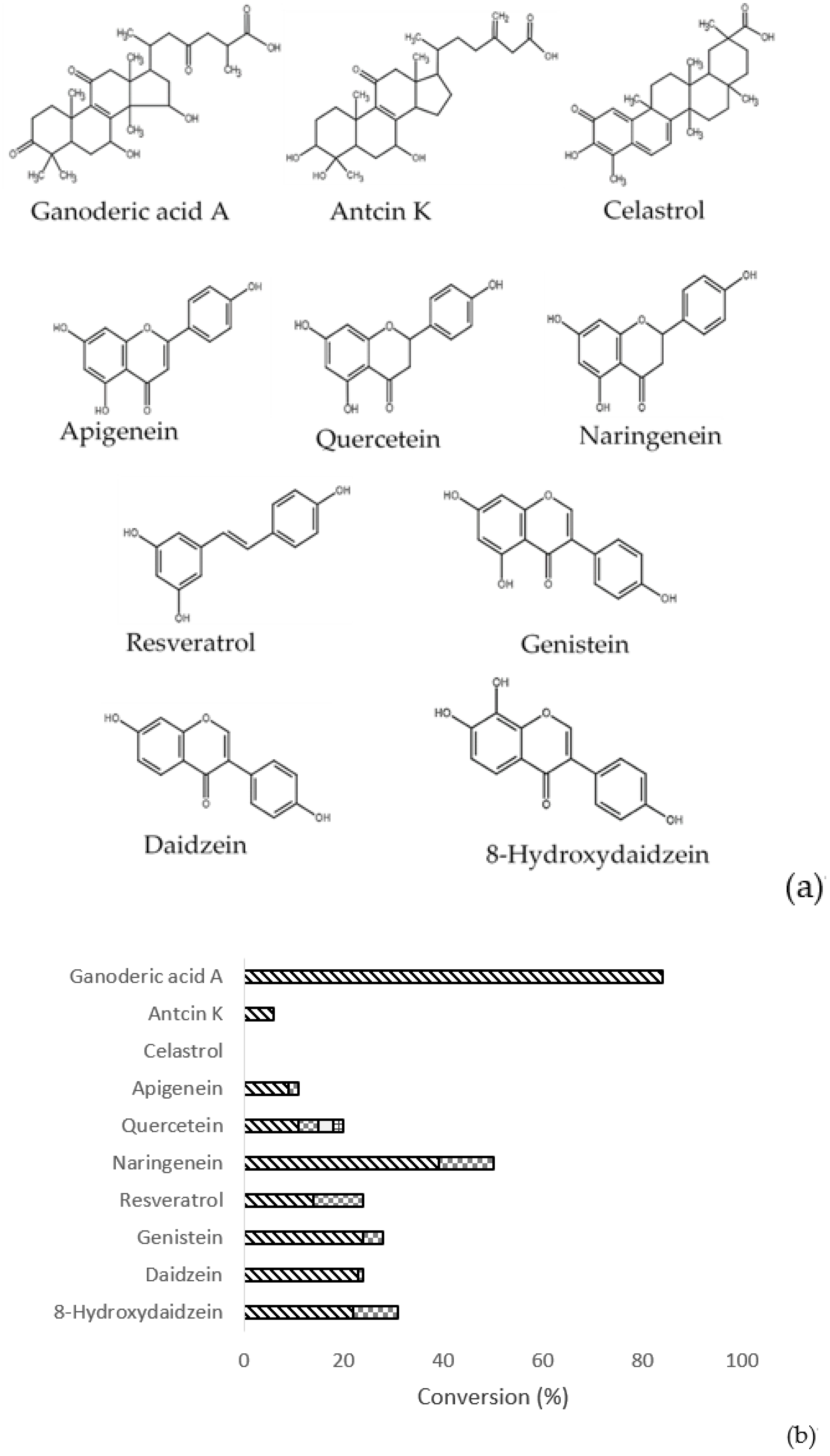
2.7. Substrate Specificity of BtGT_16345
2.8. Kinetic Study of BtGT_16345 toward GAA
3. Discussion
4. Materials and Methods
4.1. Microorganism and Chemicals
4.2. Whole-Genome Sequencing
4.3. Genome Assembly and Annotation
4.4. Reclassification of GA A07 Strain
4.5. Identification and Analysis of GT Genes
4.6. Fermentation and Biotransformation of GAA
4.7. UPLC Analysis
4.8. Expression and Purification of GT from GA A07 Strain
4.9. In Vitro Biotransformation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GAA | Ganoderic acid A |
| GT | Glycosyltransferase |
| UDP | Uridine diphosphate |
References
- Cantarel, B.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37 (Suppl. 1), D233–D238. [Google Scholar] [CrossRef]
- Hofer, B. Recent developments in the enzymatic O-glycosylation of flavonoids. Appl. Microbiol. Biotechnol. 2016, 100, 4269–4281. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 716–739. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S.; Wu, J.J.; Wang, T.Y.; Wu, K.Y.; Chiang, C.M. Uridine diphosphate-dependent glycosyltransferases from Bacillus subtilis ATCC 6633 catalyze the 15-O-glycosylation of ganoderic acid A. Int. J. Mol. Sci. 2018, 19, 3469. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Chen, X.; Jassbi, A.R.; Xiao, J. Microbial biotransformation of bioactive flavonoids. Biotechnol. Adv. 2015, 33, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.A.; Tan, H.L.; Sultan, S.; Faridz, M.A.B.M.; Shah, M.A.B.M.; Nurfazilah, S.; Hussain, M. Microbial-catalyzed biotransformation of multifunctional triterpenoids derived from phytonutrients. Int. J. Mol. Sci. 2014, 15, 12027–12060. [Google Scholar] [CrossRef] [PubMed]
- Sultana, N.; Saify, Z.S. Enzymatic biotransformation of terpenes as bioactive agents. J. Enzym. Inhib. Med. Chem. 2013, 28, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Muffler, K.; Leipold, D.; Scheller, M.C.; Haas, C.; Steingroewer, J.; Bley, T.; Neuhaus, H.E.; Mirata, M.A.; Schrader, J.; Ulber, R. Biotransformation of triterpenes. Process Biochem. 2011, 46, 1–15. [Google Scholar] [CrossRef]
- Parra, A.; Rivas, F.; Garcia-Granados, A.; Martinez, A. Microbial transformation of triterpenoids. Mini-Rev. Org. Chem. 2009, 6, 307–320. [Google Scholar] [CrossRef]
- Shimoda, K.; Hamada, H.; Hamada, H. Synthesis of xylooligosaccharides of daidzein and their anti-oxidant and anti-allergic activities. Int. J. Mol. Sci. 2011, 12, 5616–5625. [Google Scholar] [CrossRef]
- Chiang, C.M.; Wang, T.Y.; Yang, S.Y.; Wu, J.Y.; Chang, T.S. Production of new isoflavone glucosides from glycosylation of 8-hydroxydaidzein by glycosyltransferase from Bacillus subtilis ATCC 6633. Catalysts 2018, 8, 349. [Google Scholar] [CrossRef]
- Chang, T.S.; Wang, T.Y.; Yang, S.Y.; Kao, Y.H.; Wu, J.J.; Chiang, C.M. Potential industrial production of a well-soluble, alkaline-stable, and anti-inflammatory isoflavone glucoside from 8-hydroxydaidzein glucosylated by recombinant amylosucrase of Deinococcus geothermalis. Molecules 2019, 24, 2236. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Y.; Zeng, J.Z.; Wong, A.S.T. Chemical structures and pharmacological profiles of ginseng saponins. Molecules 2019, 24, 2443. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Asaka, Y. Structures of ganoderic acid A and B, two new lanostane type bitter triterpenes from Ganoderma lucidum (FR.) Karst. Helv. Chim. Acta 1982, 65, 611–619. [Google Scholar] [CrossRef]
- Xia, Q.; Zhang, H.; Sun, X.; Zhao, H.; Wu, L.; Zhu, D.; Yang, G.; Shao, Y.; Zhang, X.; Mao, X.; et al. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.W.; Zhao, W.; Zhong, J.J. Biotechnological production and application of ganoderic acids. Appl. Microbiol. Biotechnol. 2010, 87, 457–466. [Google Scholar]
- Liang, C.; Tian, D.; Liu, Y.; Li, H.; Zhu, J.; Li, M.; Xin, M.; Xia, J. Review of the molecular mechanisms of Ganoderma lucidum triterpenoids: Ganoderic acids A, C2, D, F, DM, X and Y. Eur. J. Med. Chem. 2019, 174, 130–141. [Google Scholar] [CrossRef]
- Chang, T.S.; Chiang, C.M.; Wang, T.Y.; Lee, C.H.; Lee, Y.W.; Wu, J.Y. New triterpenoid from novel triterpenoid 15-O-glycosylation on ganoderic acid A by intestinal bacteria of zebrafish. Molecules 2018, 23, 2345. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Yao, P.; Zhu, Y.; Men, Y.; Zeng, Y.; Yang, J.; Sun, Y. Exploiting the aglycon promiscuity of glycosyltransferase Bs-YjiC from Bacillus subtilis and its application in synthesis of glycosides. J. Biotechnol. 2017, 248, 69–76. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Yang, J.; Zhu, Y.; Men, Y.; Zeng, Y.; Cai, Y.; Dong, C.; Dai, Z.; Zhang, X.; et al. Use of a promiscuous glycosyltransferase from Bacillus subtilis 168 for the enzymatic synthesis of novel protopanaxtriol-type ginsenosides. J. Agric. Food Chem. 2017, 66, 943–949. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Yang, J.; Men, Y.; Zeng, Y.; Cai, Y.; Sun, Y. Enzymatic synthesis of novel glycyrrhizic acid glucosides using a promiscuous Bacillus glycosyltransferase. Catalysts 2018, 8, 615. [Google Scholar] [CrossRef]
- Li, K.; Feng, J.; Kuang, Y.; Song, W.; Zhang, M.; Ji, S.; Qiao, X.; Ye, M. Enzymatic synthesis of bufadienolide O-glycosides as potent antitumor agents using a microbial glycosyltransferase. Adv. Syn. Cat. 2017, 359, 3765–3772. [Google Scholar] [CrossRef]
- Chen, K.; He, J.; Hu, Z.; Song, W.; Yu, L.; Li, K.; Qiao, X.; Ye, M. Enzymatic glycosylation of oleanane-type triterpenoids. J. Asia. Nat. Prod. Res. 2018, 20, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Hu, Z.; Zhang, T.; Gong, T.; Chen, J.; Zhu, P.; Li, Y.; Yang, J. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab. Eng. 2017, 44, 60–69. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010, 11, 119. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Zhang, T.T.; Gong, T.; Hu, Z.F.; Gu, A.D.; Yang, J.L.; Zhu, P. Enzymatic synthesis of unnatural ginsenosides using a promiscuous UDP-glucosyltransferase from Bacillus subtilis. Molecules 2018, 23, 2797. [Google Scholar] [CrossRef]
- Wang, D.D.; Jin, Y.; Wang, C.; Kim, Y.J.; Perez, J.E.J.; Baek, N.I.; Mathiyalagan, R.; Markus, J.; Yang, D.C. Rare ginsenoside Ia synthesized from F1 by cloning and overexpression of the UDP-glycosyltransferase gene from Bacillus subtilis: Synthesis, characterization, and in vitro melanogenesis inhibition activity in BL6B16 cells. J. Gingeng Res. 2018, 42, 42–49. [Google Scholar] [CrossRef]
- Adachi, J.; Hasegawa, M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J. Mol. Evol. 1996, 42, 459–468. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 25.1–25.35. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.S.; Chiang, C.M.; Siao, Y.Y.; Wu, J.Y. Sequential biotransformation of antcin K by Bacillus subtilis ATCC 6633. Catalysts 2018, 8, 349. [Google Scholar] [CrossRef]
- Wu, S.C.; Chang, C.W.; Lin, C.W.; Hsu, Y.C. Production of 8-hydroxydaidzein polyphenol using biotransformation by Aspergillus oryzae. Food Sci. Technol. Res. 2015, 21, 557–562. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H. Fast and accurate long-read assembly with wtdbg2. bioRxiv 2019. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, 1–6. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez, R.L.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Wu, Y.W. ezTree: An automated pipeline for identifying phylogenetic marker genes and inferring evolutionary relationships among uncultivated prokaryotic draft genomes. BMC Genom. 2018, 19 (Suppl. 1), 921. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]





| GT | KM (μM) | kcat (s−1) | kcat/KM (s−1 mM−1) |
|---|---|---|---|
| BsUGT398 | 90.71 ± 14.86 | 0.1401 ± 0.0051 | 1.5445 ± 0.2592 |
| BsUGT489 | 793.96 ± 124.09 | 0.9336 ± 0.0626 | 1.1759 ± 0.2000 |
| BtGT_16345 | 263.82 ± 24.78 | 0.2944 ± 0.0109 | 1.1159 ± 0.1127 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, T.-S.; Wang, T.-Y.; Hsueh, T.-Y.; Lee, Y.-W.; Chuang, H.-M.; Cai, W.-X.; Wu, J.-Y.; Chiang, C.-M.; Wu, Y.-W. A Genome-Centric Approach Reveals a Novel Glycosyltransferase from the GA A07 Strain of Bacillus thuringiensis Responsible for Catalyzing 15-O-Glycosylation of Ganoderic Acid A. Int. J. Mol. Sci. 2019, 20, 5192. https://doi.org/10.3390/ijms20205192
Chang T-S, Wang T-Y, Hsueh T-Y, Lee Y-W, Chuang H-M, Cai W-X, Wu J-Y, Chiang C-M, Wu Y-W. A Genome-Centric Approach Reveals a Novel Glycosyltransferase from the GA A07 Strain of Bacillus thuringiensis Responsible for Catalyzing 15-O-Glycosylation of Ganoderic Acid A. International Journal of Molecular Sciences. 2019; 20(20):5192. https://doi.org/10.3390/ijms20205192
Chicago/Turabian StyleChang, Te-Sheng, Tzi-Yuan Wang, Tzu-Yu Hsueh, Yu-Wen Lee, Hsin-Mei Chuang, Wen-Xuan Cai, Jiumn-Yih Wu, Chien-Min Chiang, and Yu-Wei Wu. 2019. "A Genome-Centric Approach Reveals a Novel Glycosyltransferase from the GA A07 Strain of Bacillus thuringiensis Responsible for Catalyzing 15-O-Glycosylation of Ganoderic Acid A" International Journal of Molecular Sciences 20, no. 20: 5192. https://doi.org/10.3390/ijms20205192
APA StyleChang, T.-S., Wang, T.-Y., Hsueh, T.-Y., Lee, Y.-W., Chuang, H.-M., Cai, W.-X., Wu, J.-Y., Chiang, C.-M., & Wu, Y.-W. (2019). A Genome-Centric Approach Reveals a Novel Glycosyltransferase from the GA A07 Strain of Bacillus thuringiensis Responsible for Catalyzing 15-O-Glycosylation of Ganoderic Acid A. International Journal of Molecular Sciences, 20(20), 5192. https://doi.org/10.3390/ijms20205192







