Interleukin-6 Induces Myogenic Differentiation via JAK2-STAT3 Signaling in Mouse C2C12 Myoblast Cell Line and Primary Human Myoblasts
Abstract
:1. Introduction
2. Results
2.1. Effect of IL-6 on C2C12 Cell Cycling is Mediated via the JAK-STAT Signaling Cascade
2.2. Primary Human Myoblasts (PHM) Characterisation
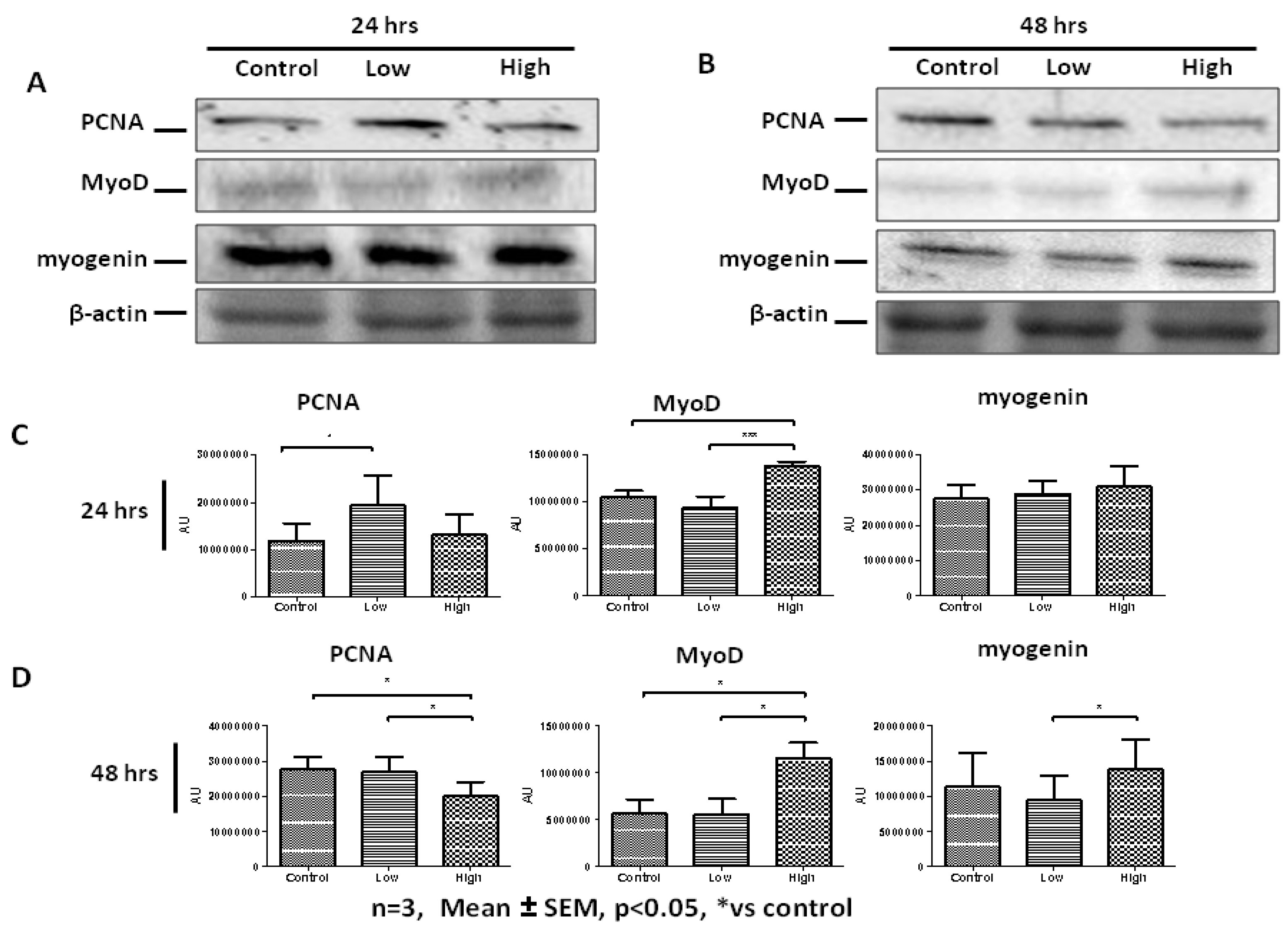
2.3. High IL-6 Concentration Induced Myogenic Differentiation in Primary Human Myoblasts (PHMs)
2.4. IL-6 Concentration-Dependent Activation of JAK2-STAT3 Signaling in Human Myogenesis
2.5. IL-6 Receptor Regulation in Response to IL-6 Treatment
3. Discussion
4. Materials and Methods
4.1. IL-6 Treatment Protocol
4.2. Cell Culture
4.3. Isolation and Culture of Human Primary Myoblasts
4.4. Human Primary Myoblasts Characterisation
4.5. Cell Cycle Analysis
4.6. Immunoblot Analysis
4.7. Immunocytochemistry Staining
4.8. RNA Preparation and RT-qPCR
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CD34 | Cluster of Differentiation 34 |
| FBS | Fetal Bovine Serum |
| HGF | Hepatocyte Growth Factor |
| IL-6 | Interleukin 6 |
| JAK | Janus kinase |
| LIF | Leukemia Inhibitory Factor |
| MEF | Myocyte Enhancer-binding Factor |
| MRFs | Myogenic Regulatory Factors |
| PCNA | Proliferating Cell Nuclear Antigen |
| PHM | Primary Human Myoblast |
| SOCS | Suppressor of Cytokine Signaling |
| STAT | Signal Transducer and Activator of Transcription |
| TGF-β | Transforming Growth Factor-β |
References
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem cell function, self-renewal and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.E.; Partridge, T.A. Muscle satellite cells. Int. J. Biochem. Cell Biol. 2003, 35, 1151–1156. [Google Scholar] [CrossRef]
- Holterman, C.E.; Rudnicki, M.A. Molecular regulation of satellite cell function. Semin. Cell Devel. Biol. 2005, 16, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Berberoglu, M.A.; Gallagher, T.L.; Morrow, Z.T.; Talbot, J.C.; Hromowyk, K.J.; Tenente, I.M.; Langenau, D.M.; Amacher, S.L. Satellite-like cells contribute to pax7-dependent skeletal muscle repair in adult zebrafish. Dev. Biol 2017, 424, 162–180. [Google Scholar] [CrossRef]
- Dumont, N.A.; Rudnicki, M.A. Characterizing satellite cells and myogenic progenitors during skeletal muscle regeneration. Methods Mol. Biol. 2017, 1560, 179–188. [Google Scholar]
- Fukada, S.; Uezumi, A.; Ikemoto, M.; Masuda, S.; Segawa, M.; Tanimura, N.; Yamamoto, H.; Miyagoe-Suzuki, Y.; Takeda, S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 2007, 25, 2448–2459. [Google Scholar] [CrossRef]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Chen, J.C.; Goldhamer, D.J. Skeletal muscle stem cells. Reprod. Biol. Endocrinol. RBE 2003, 1, 101. [Google Scholar] [CrossRef]
- Forcina, L.; Miano, C.; Pelosi, L.; Musaro, A. An Overview about the biology of skeletal muscle satellite cells. Curr. Genom. 2019, 20, 24–37. [Google Scholar] [CrossRef]
- Suzuki, T.; Takaishi, H.; Sakata, T.; Do, M.K.; Hara, M.; Sato, A.; Mizunoya, W.; Nishimura, T.; Hattori, A.; Ikeuchi, Y.; et al. In vitro measurement of post-natal changes in proliferating satellite cell frequency during rat muscle growth. Anim. Sci. J. Nihon Chikusan Gakkaiho 2010, 81, 245–251. [Google Scholar]
- Rando, T.A. Stem cells, ageing and the quest for immortality. Nature 2006, 441, 1080–1086. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.D.; Rando, T.A. Stem cell review series: Aging of the skeletal muscle stem cell niche. Aging Cell 2008, 7, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Lok, C. Cachexia: The last illness. Nat. News 2015, 528, 182. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Krauss, R.S. Muscle stem cells in developmental and regenerative myogenesis. Curr. Opin. Clin. Nutr. Metab. Care 2010, 13, 243–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuang, S.; Charge, S.B.; Seale, P.; Huh, M.; Rudnicki, M.A. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J. Cell Biol. 2006, 172, 103–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKinnell, I.W.; Ishibashi, J.; Le Grand, F.; Punch, V.G.; Addicks, G.C.; Greenblatt, J.F.; Dilworth, F.J.; Rudnicki, M.A. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008, 10, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Charge, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef] [PubMed]
- Christov, C.; Chretien, F.; Abou-Khalil, R.; Bassez, G.; Vallet, G.; Authier, F.J.; Bassaglia, Y.; Shinin, V.; Tajbakhsh, S.; Chazaud, B.; et al. Muscle satellite cells and endothelial cells: Close neighbors and privileged partners. Mol. Biol. Cell 2007, 18, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.; Kuroda, K.; Le Grand, F.; Rudnicki, M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 2007, 129, 999–1010. [Google Scholar] [CrossRef]
- Le Grand, F.; Rudnicki, M. Satellite and stem cells in muscle growth and repair. Development 2007, 134, 3953–3957. [Google Scholar] [CrossRef] [Green Version]
- Le Grand, F.; Rudnicki, M.A. Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 2007, 19, 628–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzeszczynska, J.; Meyer, A.; McGregor, R.; Schilb, A.; Degen, S.; Tadini, V.; Johns, N.; Langen, R.; Schols, A.; Glass, D.J.; et al. Alterations in the in vitro and in vivo regulation of muscle regeneration in healthy ageing and the influence of sarcopenia. J. Cachexia Sarcopenia Muscle 2018, 9, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Karalaki, M.; Fili, S.; Philippou, A.; Koutsilieris, M. Muscle regeneration: Cellular and molecular events. In Vivo 2009, 23, 779–796. [Google Scholar] [PubMed]
- Kim, J.H.; Jin, P.; Duan, R.; Chen, E.H. Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Devel. 2015, 32, 162–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackey, A.L.; Magnan, M.; Chazaud, B.; Kjaer, M. Human skeletal muscle fibroblasts stimulate in vitro myogenesis and in vivo muscle regeneration. J. Physiol. 2017, 595, 5115–5127. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Devel. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef]
- Conboy, I.M.; Conboy, M.J.; Wagers, A.J.; Girma, E.R.; Weissman, I.L.; Rando, T.A. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 2005, 433, 760–764. [Google Scholar] [CrossRef]
- Wagers, A.J.; Conboy, I.M. Cellular and molecular signatures of muscle regeneration: Current concepts and controversies in adult myogenesis. Cell 2005, 122, 659–667. [Google Scholar] [CrossRef]
- Yaffe, D.; Saxel, O. A myogenic cell line with altered serum requirements for differentiation. Differentiation 1977, 7, 159–166. [Google Scholar] [CrossRef]
- Lawson, M.A.; Purslow, P.P. Differentiation of myoblasts in serum-free media: Effects of modified media are cell line-specific. Cells Tissues Organs 2000, 167, 130–137. [Google Scholar] [CrossRef]
- Menconi, M.; Gonnella, P.; Petkova, V.; Lecker, S.; Hasselgren, P.O. Dexamethasone and corticosterone induce similar but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J. Cell. Biochem. 2008, 105, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Pittner, R.A.; Wolfe-Lopez, D.; Young, A.A.; Beaumont, K. Different pharmacological characteristics in L6 and C2C12 muscle cells and intact rat skeletal muscle for amylin, CGRP and calcitonin. Br. J. Pharmacol. 1996, 117, 847–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.M.; Sather, B.K.; Burney, E.R.; Ehrlicher, S.E.; Stierwalt, H.D.; Franco, M.C.; Newsom, S.A. Robust intrinsic differences in mitochondrial respiration and H2O2 emission between L6 and C2C12 cells. Am. J. Physiol. Cell Physiol. 2019, 317, C339–C347. [Google Scholar] [CrossRef]
- Sultan, K.R.; Henkel, B.; Terlou, M.; Haagsman, H.P. Quantification of hormone-induced atrophy of large myotubes from C2C12 and L6 cells: Atrophy-inducible and atrophy-resistant C2C12 myotubes. Am. J. Physiol. Cell Physiol. 2006, 290, C650–C659. [Google Scholar] [CrossRef] [PubMed]
- Asfour, H.A.; Allouh, M.Z.; Said, R.S. Myogenic regulatory factors: The orchestrators of myogenesis after 30 years of discovery. Exp. Biol. Med. 2018, 243, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Hernandez, J.M.; Garcia-Gonzalez, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Devel. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Devel. Biol. 2017, 72, 19–32. [Google Scholar] [CrossRef]
- Black, B.L.; Molkentin, J.D.; Olson, E.N. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell. Biol. 1998, 18, 69–77. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Olson, E.N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 1996, 93, 9366–9373. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Olson, E.N. Defining the regulatory networks for muscle development. Curr. Opin. Genet. Devel. 1996, 6, 445–453. [Google Scholar] [CrossRef]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Devel. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 2005, 132, 2685–2695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabeshima, Y.; Hanaoka, K.; Hayasaka, M.; Esumi, E.; Li, S.; Nonaka, I.; Nabeshima, Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 1993, 364, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Hasty, P.; Bradley, A.; Morris, J.H.; Edmondson, D.G.; Venuti, J.M.; Olson, E.N.; Klein, W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993, 364, 501–506. [Google Scholar] [CrossRef]
- Trenerry, M.K.; Della Gatta, P.A.; Cameron-Smith, D. JAK/STAT signaling and human in vitro myogenesis. BMC Physiol. 2011, 11, 6. [Google Scholar] [CrossRef]
- Chakravarthy, M.V.; Abraha, T.W.; Schwartz, R.J.; Fiorotto, M.L.; Booth, F.W. Insulin-like growth factor-I extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3’-kinase/Akt signaling pathway. J. Biol. Chem. 2000, 275, 35942–35952. [Google Scholar] [CrossRef]
- Mourkioti, F.; Rosenthal, N. IGF-1, inflammation and stem cells: Interactions during muscle regeneration. Trends Immunol. 2005, 26, 535–542. [Google Scholar] [CrossRef]
- Serrano, A.L.; Baeza-Raja, B.; Perdiguero, E.; Jardi, M.; Munoz-Canoves, P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008, 7, 33–44. [Google Scholar] [CrossRef]
- Spangenburg, E.E.; Booth, F.W. Multiple signaling pathways mediate LIF-induced skeletal muscle satellite cell proliferation. Am. J. Physiol. Cell Physiol. 2002, 283, C204–C211. [Google Scholar] [CrossRef] [Green Version]
- Allen, R.E.; Sheehan, S.M.; Taylor, R.G.; Kendall, T.L.; Rice, G.M. Hepatocyte growth factor activates quiescent skeletal muscle satellite cells in vitro. J. Cell Physiol. 1995, 165, 307–312. [Google Scholar] [CrossRef]
- Miller, K.J.; Thaloor, D.; Matteson, S.; Pavlath, G.K. Hepatocyte growth factor affects satellite cell activation and differentiation in regenerating skeletal muscle. Am. J. Physiol. Cell Physiol. 2000, 278, C174–C181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKay, B.R.; De Lisio, M.; Johnston, A.P.; O’Reilly, C.E.; Phillips, S.M.; Tarnopolsky, M.A.; Parise, G. Association of interleukin-6 signalling with the muscle stem cell response following muscle-lengthening contractions in humans. PLoS ONE 2009, 4, e6027. [Google Scholar] [CrossRef] [PubMed]
- Hiscock, N.; Chan, M.H.; Bisucci, T.; Darby, I.A.; Febbraio, M.A. Skeletal myocytes are a source of interleukin-6 mRNA expression and protein release during contraction: Evidence of fiber type specificity. Faseb. J. 2004, 18, 992–994. [Google Scholar] [CrossRef] [PubMed]
- Steensberg, A.; Keller, C.; Starkie, R.L.; Osada, T.; Febbraio, M.A.; Pedersen, B.K. IL-6 and TNF-alpha expression in and release from, contracting human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E1272–E1278. [Google Scholar] [CrossRef]
- Begue, G.; Douillard, A.; Galbes, O.; Rossano, B.; Vernus, B.; Candau, R.; Py, G. Early activation of rat skeletal muscle IL-6/STAT1/STAT3 dependent gene expression in resistance exercise linked to hypertrophy. PLoS ONE 2013, 8, e57141. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Lee, C.K. What does Stat3 do? J. Clin. Investig. 2002, 109, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- Trenerry, M.K.; Carey, K.A.; Ward, A.C.; Cameron-Smith, D. STAT3 signaling is activated in human skeletal muscle following acute resistance exercise. J. Appl. Physiol. 2007, 102, 1483–1489. [Google Scholar] [CrossRef] [Green Version]
- Bromberg, J.; Darnell, J.E., Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 2000, 19, 2468–2473. [Google Scholar] [CrossRef] [Green Version]
- Toth, K.G.; McKay, B.R.; De Lisio, M.; Little, J.P.; Tarnopolsky, M.A.; Parise, G. IL-6 induced STAT3 signalling is associated with the proliferation of human muscle satellite cells following acute muscle damage. PLoS ONE 2011, 6, e17392. [Google Scholar] [CrossRef]
- Ivanova, A.V.; Ivanov, S.V.; Zhang, X.; Ivanov, V.N.; Timofeeva, O.A.; Lerman, M.I. STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. J. Mol. Biol. 2004, 340, 641–653. [Google Scholar] [CrossRef]
- Cooney, R.N. Suppressors of cytokine signaling (SOCS): Inhibitors of the JAK/STAT pathway. Shock 2002, 17, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Maier, E.; Mittermeir, M.; Ess, S.; Neuper, T.; Schmiedlechner, A.; Duschl, A.; Horejs-Hoeck, J. Prerequisites for functional interleukin 31 signaling and its feedback regulation by suppressor of cytokine signaling 3 (SOCS3). J. Biol. Chem. 2015, 290, 24747–24759. [Google Scholar] [CrossRef] [PubMed]
- Hackett, A.R.; Lee, D.H.; Dawood, A.; Rodriguez, M.; Funk, L.; Tsoulfas, P.; Lee, J.K. STAT3 and SOCS3 regulate NG2 cell proliferation and differentiation after contusive spinal cord injury. Neurobiol. Dis. 2016, 89, 10–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Xi, Y.; Chen, J.; Zhu, P.; Kang, J.; Zou, Z.; Wang, F.; Bu, S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Segales, J.; Perdiguero, E.; Munoz-Canoves, P. Regulation of muscle stem cell functions: A focus on the p38 MAPK signaling pathway. Front. Cell Devel. Biol. 2016, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, J.J.; Gadina, M.; Schreiber, R.D. Cytokine signaling in 2002: New surprises in the Jak/Stat pathway. Cell 2002, 109, S121–S131. [Google Scholar] [CrossRef]
- Hirano, T. Interleukin 6 and its receptor: Ten years later. Int. Rev. Immunol. 1998, 16, 249–284. [Google Scholar] [CrossRef]
- Rose-John, S. Interleukin-6 family cytokines. Cold Spring Harb. Perspect. Biol. 2018, 10, a028415. [Google Scholar] [CrossRef]
- Rose-John, S.; Waetzig, G.H.; Scheller, J.; Grotzinger, J.; Seegert, D. The IL-6/sIL-6R complex as a novel target for therapeutic approaches. Exp. Opin. Ther. Targets 2007, 11, 613–624. [Google Scholar] [CrossRef]
- Peters, M.; Müller, A.M.; Rose-John, S. Interleukin-6 and soluble interleukin-6 receptor: Direct stimulation of gp130 and hematopoiesis. Blood 1998, 92, 3495–3504. [Google Scholar] [CrossRef]
- Rose-John, S.; Heinrich, P.C. Soluble receptors for cytokines and growth factors: Generation and biological function. Biochem. J. 1994, 300, 281. [Google Scholar] [CrossRef]
- Munoz-Canoves, P.; Scheele, C.; Pedersen, B.K.; Serrano, A.L. Interleukin-6 myokine signaling in skeletal muscle: A double-edged sword? FEBS J. 2013, 280, 4131–4148. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S.; Scheller, J.; Elson, G.; Jones, S.A. Interleukin-6 biology is coordinated by membrane-bound and soluble receptors: Role in inflammation and cancer. J. Leukoc. Biol. 2006, 80, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.N.; Baik, E.J. JAK-STAT pathway and myogenic differentiation. Jak-Stat 2013, 2, e23282. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. Publ. Protein Soc. 2018, 27, 1984–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Megeney, L.A.; Perry, R.L.; LeCouter, J.E.; Rudnicki, M.A. bFGF and LIF signaling activates STAT3 in proliferating myoblasts. Dev. Genet. 1996, 19, 139–145. [Google Scholar] [CrossRef]
- Kami, K.; Senba, E. In vivo activation of STAT3 signaling in satellite cells and myofibers in regenerating rat skeletal muscles. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2002, 50, 1579–1589. [Google Scholar] [CrossRef]
- Manabe, Y.; Fujii, N.L. Experimental research models for skeletal muscle contraction. J. Phys. Fit. Sports Med. 2016, 5, 373–377. [Google Scholar] [CrossRef] [Green Version]
- Manabe, Y.; Ogino, S.; Ito, M.; Furuichi, Y.; Takagi, M.; Yamada, M.; Goto-Inoue, N.; Ono, Y.; Fujii, N.L. Evaluation of an in vitro muscle contraction model in mouse primary cultured myotubes. Anal. Biochem. 2016, 497, 36–38. [Google Scholar] [CrossRef]
- McMahon, D.K.; Anderson, P.A.; Nassar, R.; Bunting, J.B.; Saba, Z.; Oakeley, A.E.; Malouf, N.N. C2C12 cells: Biophysical, biochemical and immunocytochemical properties. Am. J. Physiol. 1994, 266, C1795–C1802. [Google Scholar] [CrossRef]
- Brandt, A.M.; Kania, J.M.; Reinholt, B.M.; Johnson, S.E. Human IL6 stimulates bovine satellite cell proliferation through a Signal transducer and activator of transcription 3 (STAT3)-dependent mechanism. Domest. Anim. Endocrinol. 2018, 62, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Wada, E.; Tanihata, J.; Iwamura, A.; Takeda, S.; Hayashi, Y.K.; Matsuda, R. Treatment with the anti-IL-6 receptor antibody attenuates muscular dystrophy via promoting skeletal muscle regeneration in dystrophin-/utrophin-deficient mice. Skelet. Muscle 2017, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Tidball, J.G. Inflammatory processes in muscle injury and repair. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 288, R345–R353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoene, M.; Runge, H.; Haring, H.U.; Schleicher, E.D.; Weigert, C. Interleukin-6 promotes myogenic differentiation of mouse skeletal muscle cells: Role of the STAT3 pathway. Am. J. Physiol. Cell Physiol. 2013, 304, C128–C136. [Google Scholar] [CrossRef]
- Wang, K.; Wang, C.; Xiao, F.; Wang, H.; Wu, Z. JAK2/STAT2/STAT3 are required for myogenic differentiation. J. Biol. Chem. 2008, 283, 34029–34036. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Shibasaki, A.; Naka, A.; Saito, H.; Iida, K. Lactate promotes myoblast differentiation and myotube hypertrophy via a pathway involving MyoD in vitro and enhances muscle regeneration in vivo. Int. J. Mol. Sci. 2018, 19, 3649. [Google Scholar] [CrossRef]
- Sun, L.; Ma, K.; Wang, H.; Xiao, F.; Gao, Y.; Zhang, W.; Wang, K.; Gao, X.; Ip, N.; Wu, Z. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J. Cell Biol. 2007, 179, 129–138. [Google Scholar] [CrossRef]
- Jiao, Y.; Huang, B.; Chen, Y.; Hong, G.; Xu, J.; Hu, C.; Wang, C. Integrated analyses reveal overexpressed Notch1 promoting porcine satellite cells’ proliferation through regulating the cell cycle. Int. J. Mol. Sci. 2018, 19, 271. [Google Scholar] [CrossRef]
- Hassan, W.; Ding, L.; Gao, R.Y.; Liu, J.; Shang, J. Interleukin-6 signal transduction and its role in hepatic lipid metabolic disorders. Cytokine 2014, 66, 133–142. [Google Scholar] [CrossRef]
- Moresi, V.; Adamo, S.; Berghella, L. The JAK/STAT pathway in skeletal muscle pathophysiology. Front. Physiol. 2019, 10, 500. [Google Scholar] [CrossRef]
- Diao, Y.; Wang, X.; Wu, Z. SOCS1, SOCS3 and PIAS1 promote myogenic differentiation by inhibiting the leukemia inhibitory factor-induced JAK1/STAT1/STAT3 pathway. Mol. Cell. Biol. 2009, 29, 5084–5093. [Google Scholar] [CrossRef] [PubMed]
- Dodington, D.W.; Desai, H.R.; Woo, M. JAK/STAT—Emerging players in metabolism. Trends Endocrinol. Metab. TEM 2018, 29, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Lesina, M.; Kurkowski, M.U.; Ludes, K.; Rose-John, S.; Treiber, M.; Kloppel, G.; Yoshimura, A.; Reindl, W.; Sipos, B.; Akira, S.; et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 2011, 19, 456–469. [Google Scholar] [CrossRef]
- Guadagnin, E.; Mazala, D.; Chen, Y.W. STAT3 in skeletal muscle function and disorders. Int. J. Mol. Sci. 2018, 19, 2265. [Google Scholar] [CrossRef]
- Yu, T.; Zuo, Y.; Cai, R.; Huang, X.; Wu, S.; Zhang, C.; Chin, Y.E.; Li, D.; Zhang, Z.; Xia, N.; et al. SENP1 regulates IFN-gamma-STAT1 signaling through STAT3-SOCS3 negative feedback loop. J. Mol. Cell Biol. 2017, 9, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Tierney, M.T.; Aydogdu, T.; Sala, D.; Malecova, B.; Gatto, S.; Puri, P.L.; Latella, L.; Sacco, A. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 2014, 20, 1182–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurosaka, M.; Machida, S. Interleukin-6-induced satellite cell proliferation is regulated by induction of the JAK2/STAT3 signalling pathway through cyclin D1 targeting. Cell Prolif. 2013, 46, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Della Gatta, P.A.; Cameron-Smith, D.; Peake, J.M. Acute resistance exercise increases the expression of chemotactic factors within skeletal muscle. Eur. J. Appl. Physiol. 2014, 114, 2157–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKay, B.R.; Ogborn, D.I.; Baker, J.M.; Toth, K.G.; Tarnopolsky, M.A.; Parise, G. Elevated SOCS3 and altered IL-6 signaling is associated with age-related human muscle stem cell dysfunction. Am. J. Physiol. Cell Physiol. 2013, 304, C717–C728. [Google Scholar] [CrossRef] [Green Version]
- Spangenburg, E.E. SOCS-3 induces myoblast differentiation. J. Biol. Chem. 2005, 280, 10749–10758. [Google Scholar] [CrossRef]
- Caldow, M.K.; Steinberg, G.R.; Cameron-Smith, D. Impact of SOCS3 overexpression on human skeletal muscle development in vitro. Cytokine 2011, 55, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.; Huang, X.Y.; Zhang, J.J. Identification of novel direct Stat3 target genes for control of growth and differentiation. J. Biol. Chem. 2008, 283, 3791–3798. [Google Scholar] [CrossRef] [PubMed]
- Senn, J.J.; Klover, P.J.; Nowak, I.A.; Zimmers, T.A.; Koniaris, L.G.; Furlanetto, R.W.; Mooney, R.A. Suppressor of cytokine signaling-3 (SOCS-3), a potential mediator of interleukin-6-dependent insulin resistance in hepatocytes. J. Biol. Chem. 2003, 278, 13740–13746. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, U.R.; Agergaard, J.; Couppe, C.; Grosset, J.F.; Karlsen, A.; Magnusson, S.P.; Schjerling, P.; Kjaer, M.; Mackey, A.L. Skeletal muscle morphology and regulatory signalling in endurance-trained and sedentary individuals: The influence of ageing. Exp. Gerontol. 2017, 93, 54–67. [Google Scholar] [CrossRef]
- Aoyagi, T.; Terracina, K.P.; Raza, A.; Matsubara, H.; Takabe, K. Cancer cachexia, mechanism and treatment. World J. Gastrointest. Oncol. 2015, 7, 17–29. [Google Scholar] [CrossRef]
- Baltgalvis, K.A.; Berger, F.G.; Pena, M.M.; Davis, J.M.; Muga, S.J.; Carson, J.A. Interleukin-6 and cachexia in ApcMin/+ mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R393–R401. [Google Scholar] [CrossRef]
- Okada, S.; Okusaka, T.; Ishii, H.; Kyogoku, A.; Yoshimori, M.; Kajimura, N.; Yamaguchi, K.; Kakizoe, T. Elevated serum interleukin-6 levels in patients with pancreatic cancer. Jpn. J. Clin. Oncol. 1998, 28, 12–15. [Google Scholar] [CrossRef]
- Srdic, D.; Plestina, S.; Sverko-Peternac, A.; Nikolac, N.; Simundic, A.M.; Samarzija, M. Cancer cachexia, sarcopenia and biochemical markers in patients with advanced non-small cell lung cancer-chemotherapy toxicity and prognostic value. Support. Care Cancer Off. J. Mult. Assoc. Support. Care Cancer 2016, 24, 4495–4502. [Google Scholar] [CrossRef]
- Laumonier, T.; Koenig, S.; Sauc, S.; Frieden, M. Isolation of human myoblasts, assessment of myogenic differentiation and store-operated calcium entry measurement. J. Vis. Exp. JoVE 2017, 125, e55918. [Google Scholar] [CrossRef]
- Spinazzola, J.M.; Gussoni, E. Isolation of primary human skeletal muscle cells. Bio-Protocol 2017, 7. [Google Scholar] [CrossRef]
- Blau, H.M.; Webster, C. Isolation and characterization of human muscle cells. Proc. Natl. Acad. Sci. USA 1981, 78, 5623–5627. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]















| 1°Antibody | Manufacturer | Species | Catalogue Number |
|---|---|---|---|
| MyoD | Santa Cruz | Mouse | SC32758 |
| Myogenin | Santa Cruz | Mouse | SC12732 |
| PCNA | Santa Cruz | Rabbit | SC7907 |
| pSTAT3 | Cell Signaling | Rabbit | 9131 |
| tSTAT3 | Cell Signaling | Rabbit | 9132 |
| pJAK1 | Santa Cruz | Goat | SC16773 |
| pJAK2 | Santa Cruz | Goat | SC21870 |
| SOCS3 | Santa Cruz | Rabbit | SC9023 |
| SOCS1 | Santa Cruz | Rabbit | SC9021 |
| IL-6Rα | Santa Cruz | Rabbit | SC13947 |
| β-actin | Cell Signaling | Rabbit | 4967 |
| 2°Antibody | Manufacturer | Description |
|---|---|---|
| Anti-mouse | Abcam | Goat HRP conjugated |
| Anti-rabbit | Santa Cruz | Goat HRP conjugated |
| Anti-goat | Abcam | Sheep HRP conjugated |
| 2°Antibody | Manufacturer | Species | Product Number |
|---|---|---|---|
| Alexa Fluor 488 | Invitrogen | Donkey Anti-mouse IgG | A21202 |
| Alex Fluor 546 | Invitrogen | Donkey Anti-goat IgG | A11056 |
| Alexa Fluor 594 | Invitrogen | Donkey Anti-rabbit | A21207 |
| Primer | Manufacturer | Primer Code |
|---|---|---|
| IL-6R | Applied Biosystems | Hs1075666_m1 |
| Housekeeping (B2M) | Applied Biosystems | Hs00984230_m1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steyn, P.J.; Dzobo, K.; Smith, R.I.; Myburgh, K.H. Interleukin-6 Induces Myogenic Differentiation via JAK2-STAT3 Signaling in Mouse C2C12 Myoblast Cell Line and Primary Human Myoblasts. Int. J. Mol. Sci. 2019, 20, 5273. https://doi.org/10.3390/ijms20215273
Steyn PJ, Dzobo K, Smith RI, Myburgh KH. Interleukin-6 Induces Myogenic Differentiation via JAK2-STAT3 Signaling in Mouse C2C12 Myoblast Cell Line and Primary Human Myoblasts. International Journal of Molecular Sciences. 2019; 20(21):5273. https://doi.org/10.3390/ijms20215273
Chicago/Turabian StyleSteyn, Paul J., Kevin Dzobo, Robert I. Smith, and Kathryn H. Myburgh. 2019. "Interleukin-6 Induces Myogenic Differentiation via JAK2-STAT3 Signaling in Mouse C2C12 Myoblast Cell Line and Primary Human Myoblasts" International Journal of Molecular Sciences 20, no. 21: 5273. https://doi.org/10.3390/ijms20215273





