Expression of A New Endogenous Retrovirus-Associated Transcript in Hodgkin Lymphoma Cells
Abstract
:1. Introduction
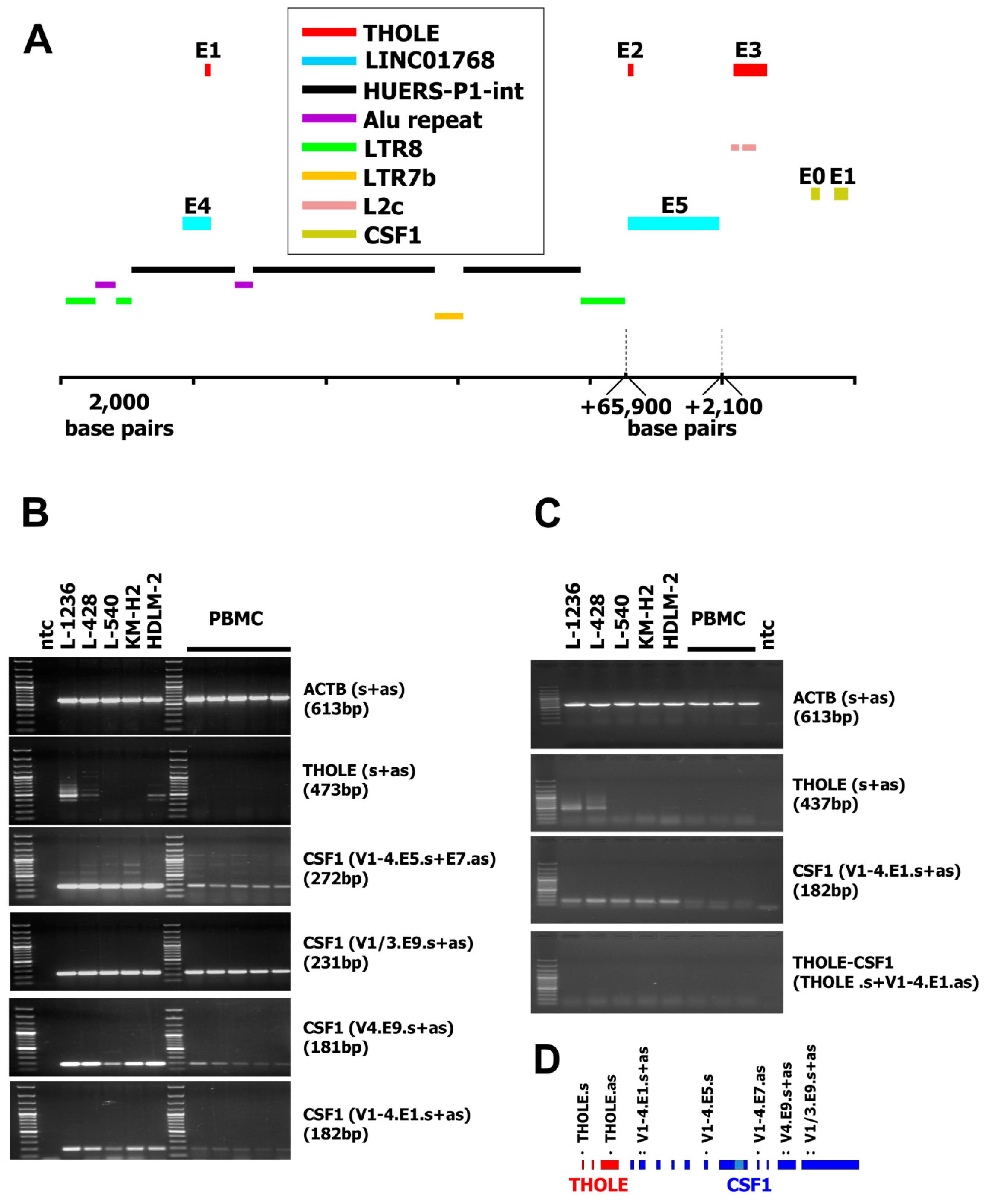
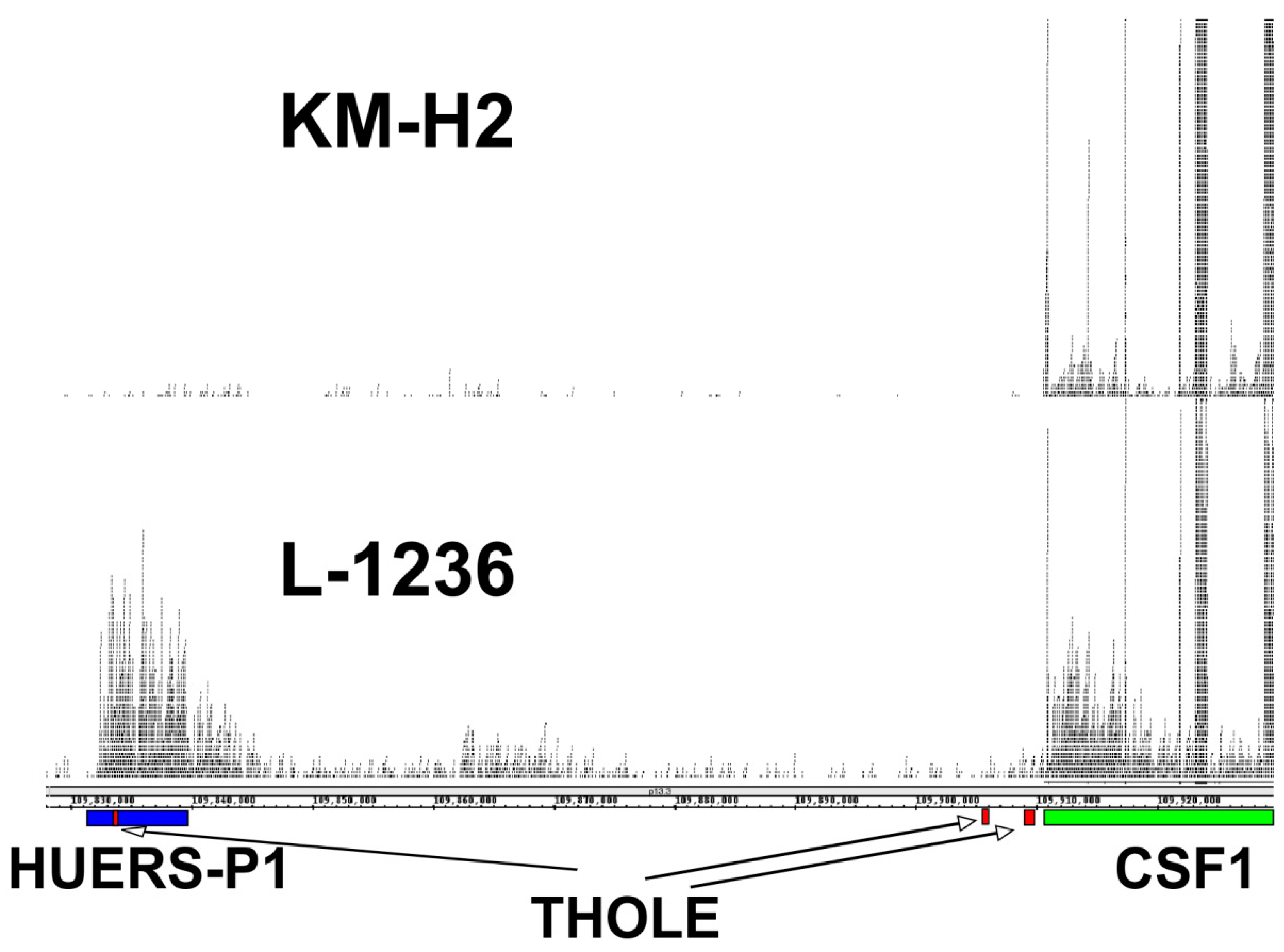
2. Results and Discussion
3. Materials and Methods
3.1. Cells and Cell Lines
3.2. DNA Isolation, RNA, RNA Isolation and Polymerase Chain Reaction (PCR)
3.3. Isolation of A THOLE cDNA–Containing Vector
3.4. Bioinformatics Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Collins, M.K. Species specificity of interleukin 2 binding to individual receptor components. Eur. J. Immunol. 1989, 19, 1517–1520. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, T.R.; Yokota, T.; Kastelein, R.; Zurawski, S.M.; Arai, N.; Takebe, Y. Species-specificity of T cell stimulating activities of IL 2 and BSF-1 (IL 4): Comparison of normal and recombinant, mouse and human IL 2 and BSF-1 (IL 4). J. Immunol. 1987, 138, 1813–1816. [Google Scholar] [PubMed]
- Balint, E.; Manolescu, N. Hodgkin’s malignant lymphoma in dogs. Lucr. Stiint. Med. Vet. Timarosa 2010, 43, 373–378. [Google Scholar]
- Walton, R.M.; Hendrick, M.J. Feline Hodgkin’s-like lymphoma: 20 cases (1992–1999). Vet. Pathol. 2001, 38, 504–511. [Google Scholar] [CrossRef]
- Durham, A.C.; Pillitteri, C.A.; San Myint, M.; Valli, V.E. Two hundred three cases of equine lymphoma classified according to the World Health Organization (WHO) classification criteria. Vet. Pathol. 2013, 50, 86–93. [Google Scholar] [CrossRef]
- Murray, P.; Bell, A. Contribution of the Epstein-Barr Virus to the pathogenesis of Hodgkin lymphoma. Curr. Top. Microbiol. Immunol. 2015, 390, 287–313. [Google Scholar]
- Vrzalikova, K.; Sunmonu, T.; Reynolds, G.; Murray, P. Contribution of Epstein-Barr Virus latent proteins to the pathogenesis of classical Hodgkin lymphoma. Pathogens 2018, 7, 59. [Google Scholar] [CrossRef]
- Lee, E.K.; Joo, E.H.; Song, K.A.; Choi, B.; Kim, M.; Kim, S.H.; Kim, S.J.; Kang, M.S. Effects of lymphocyte profile on development of EBV-induced lymphoma subtypes in humanized mice. Proc. Natl. Acad. Sci. USA 2015, 112, 13081–13086. [Google Scholar] [CrossRef] [Green Version]
- Raffegerst, S.H.; Hoelzlwimmer, G.; Kunder, S.; Mysliwietz, J.; Quintanilla-Martinez, L.; Schendel, D.J. Diverse hematological malignancies including hodgkin-like lymphomas develop in chimeric MHC class II transgenic mice. PLoS ONE 2009, 4, e8539. [Google Scholar] [CrossRef]
- Kumar, R.K. Hodgkin’s disease. SJL/J murine lymphoma. Am. J. Pathol. 1983, 110, 393–396. [Google Scholar]
- Katz, J.; Bonavida, B. Expression of superantigen-like specificities on murine sjl/j-B lymphomas—Antitumor v-Beta-17a+ T-lymphocytes use a diverse set of T-cell receptor v-alpha-chain gene-sequences. Int. J. Oncol. 1994, 4, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Waugh, E.M.; Gallagher, A.; McAulay, K.A.; Henriques, J.; Alves, M.; Bell, A.J.; Morris, J.S.; Jarrett, R.F. Gammaherpesviruses and canine lymphoma: No evidence for direct involvement in commonly occurring lymphomas. J. Gen. Virol. 2015, 96, 1863–1872. [Google Scholar] [CrossRef] [PubMed]
- Eberle, F.C.; Mani, H.; Jaffe, E.S. Histopathology of Hodgkin’s lymphoma. Cancer J. 2009, 15, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Staege, M.S. A multi-component model of Hodgkin’s lymphoma. PLoS ONE 2015, 10, e0124614. [Google Scholar] [CrossRef]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef]
- Babaian, A.; Romanish, M.T.; Gagnier, L.; Kuo, L.Y.; Karimi, M.M.; Steidl, C.; Mager, D.L. Onco-exaptation of an endogenous retroviral LTR drives IRF5 expression in Hodgkin lymphoma. Oncogene 2016, 35, 2542–2546. [Google Scholar] [CrossRef]
- Kruse, K.; Nettling, M.; Wappler, N.; Emmer, A.; Kornhuber, M.; Staege, M.S.; Grosse, I. WebHERV: A web server for the computational investigation of gene expression associated with endogenous retrovirus-like sequences. Front. Microbiol. 2018, 9, 2384. [Google Scholar] [CrossRef]
- Barth, M.; Gröger, V.; Cynis, H.; Staege, M.S. Identification of human endogenous retrovirus transcripts in Hodgkin Lymphoma cells. Mol. Biol. Rep. 2019, 46, 1885–1893. [Google Scholar] [CrossRef]
- Kowalska, M.; Tajer, J.; Chechlinska, M.; Fuksiewicz, M.; Kotowicz, B.; Kaminska, J.; Walewski, J. Serum macrophage colony-stimulating factor (M-CSF) in patients with Hodgkin lymphoma. Med. Oncol. 2012, 29, 2143–2147. [Google Scholar] [CrossRef]
- Staege, M.S.; Emmer, A. Editorial: Endogenous viral elements-links between autoimmunity and cancer? Front. Microbiol. 2018, 9, 3171. [Google Scholar] [CrossRef]
- Thomas, J.; Perron, H.; Feschotte, C. Variation in proviral content among human genomes mediated by LTR recombination. Mob. DNA 2018, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Staege, M.S.; Müller, K.; Kewitz, S.; Volkmer, I.; Mauz-Körholz, C.; Bernig, T.; Körholz, D. Expression of dual-specificity phosphatase 5 pseudogene 1 (DUSP5P1) in tumor cells. PLoS ONE 2014, 9, e89577. [Google Scholar] [CrossRef] [PubMed]
- Tomita, N.; Horii, A.; Doi, S.; Yokouchi, H.; Ogawa, M.; Mori, T.; Matsubara, K. Transcription of human endogenous retroviral long terminal repeat (LTR) sequence in a lung cancer cell line. Biochem. Biophys. Res. Commun. 1990, 166, 1–10. [Google Scholar] [CrossRef]
- Gao, C.; Xiao, M.; Ren, X.; Hayward, A.; Yin, J.; Wu, L.; Fu, D.; Li, J. Characterization and functional annotation of nested transposable elements in eukaryotic genomes. Genomics 2012, 100, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Harada, F.; Tsukada, N.; Kato, N. Isolation of three kinds of human endogenous retrovirus-like sequences using tRNA(Pro) as a probe. Nucleic Acids Res. 1987, 15, 9153–9162. [Google Scholar] [CrossRef]
- Leung, A.; Trac, C.; Kato, H.; Costello, K.R.; Chen, Z.; Natarajan, R.; Schones, D.E. LTRs activated by Epstein-Barr virus-induced transformation of B cells alter the transcriptome. Genome Res. 2018, 28, 1791–1798. [Google Scholar] [CrossRef]
- Niedobitek, G.; Päzolt, D.; Teichmann, M.; Devergne, O. Frequent expression of the Epstein-Barr virus (EBV)-induced gene, EBI3, an IL-12 p40-related cytokine, in Hodgkin and Reed-Sternberg cells. J. Pathol. 2002, 198, 310–316. [Google Scholar] [CrossRef]
- Foell, J.L.; Max, D.; Giersberg, C.; Korholz, D.; Staege, M.S. Sensitivity of Hodgkin’s lymphoma cell lines to the cell cycle inhibitor roscovitine. Anticancer Res. 2008, 28, 887–894. [Google Scholar]
- Liu, M.; Thomas, S.L.; DeWitt, A.K.; Zhou, W.; Madaj, Z.B.; Ohtani, H.; Baylin, S.B.; Liang, G.; Jones, P.A. Dual inhibition of DNA and histone methyltransferases increases viral mimicry in ovarian cancer cells. Cancer Res. 2018, 78, 5754–5766. [Google Scholar] [CrossRef]
- Bustamante Rivera, Y.Y.; Brütting, C.; Schmidt, C.; Volkmer, I.; Staege, M.S. Endogenous retrovirus 3—History, physiology, and pathology. Front. Microbiol. 2018, 8, 2691. [Google Scholar] [CrossRef]
- Fischle, W.; Wang, Y.; Allis, C.D. Binary switches and modification cassettes in histone biology and beyond. Nature 2003, 425, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Verma, U.N.; Prajapati, S.; Kwak, Y.T.; Gaynor, R.B. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 2003, 423, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Bargou, R.C.; Emmerich, F.; Krappmann, D.; Bommert, K.; Mapara, M.Y.; Arnold, W.; Royer, H.D.; Grinstein, E.; Greiner, A.; Scheidereit, C.; et al. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells. J. Clin. Investig. 1997, 100, 2961–2969. [Google Scholar] [CrossRef] [PubMed]
- Edginton-White, B.; Cauchy, P.; Assi, S.A.; Hartmann, S.; Riggs, A.G.; Mathas, S.; Cockerill, P.N.; Bonifer, C. Global long terminal repeat activation participates in establishing the unique gene expression programme of classical Hodgkin lymphoma. Leukemia 2019, 33, 1463–1474. [Google Scholar] [CrossRef]
- Gao, S.; Song, L.; Li, J.; Zhang, Z.; Peng, H.; Jiang, W.; Wang, Q.; Kang, T.; Chen, S.; Huang, W. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell. Microbiol. 2012, 14, 1849–1866. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Mueller, T.; Hantsch, C.; Volkmer, I.; Staege, M.S. Differentiation-Dependent regulation of human endogenous retrovirus K sequences and neighboring genes in germ cell tumor cells. Front. Microbiol. 2018, 9, 1253. [Google Scholar] [CrossRef]
- Staege, M.S.; Banning-Eichenseer, U.; Weissflog, G.; Volkmer, I.; Burdach, S.; Richter, G.; Mauz-Körholz, C.; Föll, J.; Körholz, D. Gene expression profiles of Hodgkin’s lymphoma cell lines with different sensitivity to cytotoxic drugs. Exp. Hematol. 2008, 36, 886–896. [Google Scholar] [CrossRef]
- Jacques, C.; Renema, N.; Lezot, F.; Ory, B.; Walkley, C.R.; Grigoriadis, A.E.; Heymann, D. Small animal models for the study of bone sarcoma pathogenesis: Characteristics, therapeutic interests and limitations. J. Bone Oncol. 2018, 12, 7–13. [Google Scholar] [CrossRef]
- Abedi, G.; Hesaraki, S.; Yadegar, O. Mandibular primitive neuroectodermal tumor in an adult dog. Iran. J. Vet. Res. 2013, 14, 261–263. [Google Scholar]
- De Cock, H.E.; Busch, M.D.; Fry, M.M.; Mehl, M.; Bollen, A.W.; Higgins, R.J. A peripheral primitive neuroectodermal tumor with generalized bone metastases in a puppy. Vet. Pathol. 2004, 41, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Hosokava, S.; Suzuki, S.; Hibino, N.; Fukuta, T.; Imai, T.; Hayakawa, K.; Nakanowatari, J.; Sagami, F. Peripheral primitive neuroectodermal tumor (peripheral neuro-epithelioma) in a dog. J. Am. Assoc. Lab. Anim. Sci. 1998, 37, 66–69. [Google Scholar]
- Lucas, M.N.; Nguyen, F.; Abadie, J.; Kane, Y.; Cuillière, P.; Wyers, M. Cerebral primitive neuroectodermal tumour in a heifer. J. Comp. Pathol. 2003, 128, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Facemire, P.R.; Facemire, L.M.; Honnold, S.P. Peripheral primitive neuroectodermal tumor in a two-year-old paint horse. J. Vet. Diagn. Investig. 2012, 24, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.; Walz, P.H. Peripheral primitive neuroectodermal tumour in a lumbar vertebra and the liver of a dromedary camel (Camelus dromedarius). J. Comp. Pathol. 2009, 141, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Long, P.H.; Schulman, F.Y.; Koestner, A.; Fix, A.S.; Campbell, M.K.; Cameron, K.N. Primitive neuroectodermal tumor in a two month-old black and white Colobus monkey. Vet. Pathol. 1998, 35, 64–67. [Google Scholar] [CrossRef]
- Minas, T.Z.; Surdez, D.; Javaheri, T.; Tanaka, M.; Howarth, M.; Kang, H.J.; Han, J.; Han, Z.Y.; Sax, B.; Kream, B.E.; et al. Combined experience of six independent laboratories attempting to create an Ewing sarcoma mouse model. Oncotarget 2017, 8, 34141–34163. [Google Scholar] [CrossRef]
- Torchia, E.C.; Boyd, K.; Rehg, J.E.; Qu, C.; Baker, S.J. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol. Cell. Biol. 2007, 27, 7918–7934. [Google Scholar] [CrossRef]
- Foell, J.L.; Volkmer, I.; Giersberg, C.; Kornhuber, M.; Horneff, G.; Staege, M.S. Loss of detectability of Charcot-Leyden crystal protein transcripts in blood cells after treatment with dimethyl sulfoxide. J. Immunol. Methods 2008, 339, 99–103. [Google Scholar] [CrossRef]
- Hoennscheidt, C.; Max, D.; Richter, N.; Staege, M.S. Expression of CD4 on Epstein-Barr virus-immortalized B cells. Scand. J. Immunol. 2009, 70, 216–225. [Google Scholar] [CrossRef]
- Gilbert, F.; Balaban-Malenbaumm, G. Genetic regulation of malignancy in human neuroblastoma cell hybrids. In Advances in Neuroblastoma Research; Evans, A.E., Ed.; Raven: New York, NY, USA, 1980; pp. 59–72. [Google Scholar]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Biedler, J.L.; Helson, L.; Spengler, B.A. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973, 33, 2643–2652. [Google Scholar] [PubMed]
- Schwab, M.; Alitalo, K.; Klempnauer, K.H.; Varmus, H.E.; Bishop, J.M.; Gilbert, F.; Brodeur, G.; Goldstein, M.; Trent, J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 1983, 305, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.; Klein, G.; Nadkarni, J.S.; Nadkarni, J.J.; Wigzell, H.; Clifford, P. Surface IgM-kappa specificity on a Burkitt lymphoma cell in vivo and in derived culture lines. Cancer Res. 1968, 28, 1300–1310. [Google Scholar] [PubMed]
- Hurwitz, R.; Hozier, J.; LeBien, T.; Minowada, J.; Gajl-Peczalska, K.; Kubonishi, I.; Kersey, J. Characterization of a leukemic cell line of the pre-B phenotype. Int. J. Cancer 1979, 23, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Schneider, U.; Schwenk, H.U.; Bornkamm, G. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer 1977, 19, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Kapp, U.; Bohlen, H.; Kornacker, M.; Schoch, C.; Stahl, B.; Mücke, S.; von Kalle, C.; Fonatsch, C.; Schaefer, H.E.; et al. Peripheral blood mononuclear cells of a patient with advanced Hodgkin’s lymphoma give rise to permanently growing Hodgkin-Reed Sternberg cells. Blood 1996, 87, 3418–3428. [Google Scholar] [CrossRef]
- Schaadt, M.; Fonatsch, C.; Kirchner, H.; Diehl, V. Establishment of a malignant, Epstein-Barr-virus (EBV)-negative cell-line from the pleura effusion of a patient with Hodgkin’s disease. Ann. Hematol. 1979, 38, 185–190. [Google Scholar] [CrossRef]
- Diehl, V.; Kirchner, H.H.; Schaadt, M.; Fonatsch, C.; Stein, H.; Gerdes, J.; Boie, C. Hodgkin’s disease: Establishment and characterization of four in vitro cell lines. J. Cancer Res. Clin. Oncol. 1981, 101, 111–124. [Google Scholar] [CrossRef]
- Kamesaki, H.; Fukuhara, S.; Tatsumi, E.; Uchino, H.; Yamabe, H.; Miwa, H.; Shirakawa, S.; Hatanaka, M.; Honjo, T. Cytochemical, immunologic, chromosomal, and molecular genetic analysis of a novel cell line derived from Hodgkin’s disease. Blood 1986, 68, 285–292. [Google Scholar] [CrossRef]
- Drexler, H.G.; Gaedicke, G.; Lok, M.S.; Diehl, V.; Minowada, J. Hodgkin’s disease derived cell lines HDLM-2 and L-428: Comparison of morphology, immunological and isoenzyme profiles. Leuk. Res. 1986, 10, 487–500. [Google Scholar] [CrossRef]
- Kempkes, B.; Spitkovsky, D.; Jansen-Dürr, P.; Ellwart, J.W.; Kremmer, E.; Delecluse, H.J.; Rottenberger, C.; Bornkamm, G.W.; Hammerschmidt, W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995, 14, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Andrews, P.W.; Damjanov, I.; Simon, D.; Banting, G.S.; Carlin, C.; Dracopoli, N.C.; Føgh, J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Investig. 1984, 50, 147–162. [Google Scholar] [PubMed]
- Brütting, C.; Narasimhan, H.; Hoffmann, F.; Kornhuber, M.E.; Staege, M.S.; Emmer, A. Investigation of endogenous retrovirus sequences in the neighborhood of genes up-regulated in a neuroblastoma model after treatment with hypoxia-mimetic cobalt chloride. Front. Microbiol. 2018, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Freese, N.H.; Norris, D.C.; Loraine, A.E. Integrated genome browser: Visual analytics platform for genomics. Bioinformatics 2016, 32, 2089–2095. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schneider, J.; Volkmer, I.; Engel, K.; Emmer, A.; Staege, M.S. Expression of A New Endogenous Retrovirus-Associated Transcript in Hodgkin Lymphoma Cells. Int. J. Mol. Sci. 2019, 20, 5320. https://doi.org/10.3390/ijms20215320
Schneider J, Volkmer I, Engel K, Emmer A, Staege MS. Expression of A New Endogenous Retrovirus-Associated Transcript in Hodgkin Lymphoma Cells. International Journal of Molecular Sciences. 2019; 20(21):5320. https://doi.org/10.3390/ijms20215320
Chicago/Turabian StyleSchneider, Jana, Ines Volkmer, Kristina Engel, Alexander Emmer, and Martin S. Staege. 2019. "Expression of A New Endogenous Retrovirus-Associated Transcript in Hodgkin Lymphoma Cells" International Journal of Molecular Sciences 20, no. 21: 5320. https://doi.org/10.3390/ijms20215320
APA StyleSchneider, J., Volkmer, I., Engel, K., Emmer, A., & Staege, M. S. (2019). Expression of A New Endogenous Retrovirus-Associated Transcript in Hodgkin Lymphoma Cells. International Journal of Molecular Sciences, 20(21), 5320. https://doi.org/10.3390/ijms20215320





