Abstract
The WRKY transcription factors are one of the most important plant-specific transcription factors and play vital roles in various biological processes. However, the functions of WRKY genes in wintersweet (Chimonanthus praecox) are still unknown. In this report, a group IIc WRKY gene, CpWRKY71, was isolated from wintersweet. CpWRKY71 was localized to the nucleus and possessed transcriptional activation activity. qRT-PCR (quantitative real-time PCR) analysis showed that CpWRKY71 was expressed in all tissues tested, with higher expression in flowers and senescing leaves. During the flower development, the highest expression was detected in the early-withering stage, an obvious expression of CpWRKY71 was also observed in the flower primordia differentiation and the bloom stage. Meanwhile, the expression of CpWRKY71 was influenced by various abiotic stress and hormone treatments. The expression patterns of the CpWRKY71 gene were further confirmed in CpWRKY71pro:GUS (β-glucuronidase) plants. Heterologous overexpression of CpWRKY71 in Arabidopsis caused early flowering. Consistent with the early flowering phenotype, the expression of floral pathway integrators and floral meristem identity (FMI) genes were significantly up-regulated in transgenic plants. In addition, we also observed that the transgenic plants of CpWRKY71 exhibited precocious leaf senescence. In conclusion, our results suggested that CpWRKY71 may be involved in the regulation of flowering and leaf senescence in Arabidopsis. Our study provides a foundation for further characterization of CpWRKY genes function in wintersweet, and also enrich our knowledge of molecular mechanism about flowering and senescence in wintersweet.
1. Introduction
Wintersweet (Chimonanthus praecox), a perennial deciduous shrub of the Calycanthaceae, is native to China and widely distributing in south-central and southwest China [1]. Wintersweet, with its beautiful color, sweet fragrance, and long flowering period (from November to March), serves as an important winter-season ornamental tree in China and is widely used as landscape plants, pot plants and cut flowers [2]. Due to its unique flowering time, some researchers have paid attention to wintersweet flower development. To date, some progress has been made in this area. For instance, the wintersweet APETALA3 (CpAP3) gene was found to partially rescue stamen development in the Arabidopsis ap3 mutant, and caused numerous morphological changes and homeotic conversions to flowers [3]. Wintersweet AGAMOUS-like6 (CpAGL6) gene was reported to promote flowering through inhibition of FLOWERING LOCUS C (FLC) and activation of APETALA1 (AP1) and FLOWERING LOCUS T (FT) and cause abnormal stamen and carpel development in later developing flowers [4]. CpCZF1 and CpCZF2, two C3H type zinc finger protein genes of wintersweet, were revealed to affect stamen development and caused the formation of abnormal flowers in transgenic Arabidopsis plants [5]. Additionally, for ornamental plants and cut flowers, the research on senescence is always one of the hot issues, which is closely related to its commercial value and market competitiveness [6]. In recent years, Sui et al. [7] reported the effects of hormones on the wintersweet cut flower senescence. However, little information is available regarding the molecular mechanisms of wintersweet senescence. According to previous research, plant growth and development processes, such as seed germination, secondary wall formation, flowering, and senescence, are regulated by a complex genetic network, in which transcriptional modulation is a crucial aspect of the complex genetic networks [8,9,10].
In the past decades, a growing number of transcription factor genes have been isolated and proved to widely participate in various developmental and physiological processes. Among them, the WRKY transcription factor family is one of the largest plant-specific transcription factor families. They are named after the conserved motif WRKYGQK and contain one or two conserved WRKY domains which are comprised of 60 amino acids at the N-terminal end and a zinc finger-like motif either C2H2 or C2HC at the C-terminal. Based on the number of WRKY domains and the type of zinc-finger motif, the family can be classified into three subfamilies, namely, group І (two WRKY domains and C2H2 zinc-finger), group ІІ (one WRKY domain and C2H2 zinc-finger) and group Ш (one WRKY domain and C2HC zinc-finger). Additionally, based on additional structural motifs outside the WRKY domain, group ІІ can be further divided into five subgroups (IIa, IIb, IIc, IId, and IIe). WRKY proteins regulate the expression of downstream genes by recognizing and binding to the W-box (TTGACC/T) present in their promoters [11]. Since SPF1 (the first WRKY gene) was isolated from sweet potato [12], a large number of WRKY genes have been found subsequently in various plants [13,14,15,16,17,18]. According to previous reports, WRKY proteins are widely involved in plant developmental and physiological processes, and various biotic and abiotic stress responses. For example, Arabidopsis WRKY41 was reported to control Arabidopsis seed dormancy by directly regulating the expression of ABI3 [19]. Overexpression of cotton GhWRKY17 accelerated leaf senescence in A. thaliana [20]. Dlf1, a WRKY transcription factor gene which was isolated from rice, was reported to participate in the regulation of flowering time and plant height in rice [21]. Overexpression of Chrysanthemum CmWRKY48 enhanced aphid resistance of transgenic chrysanthemum [22]. Overexpression of wheat TaWRKY93 in A. thaliana enhanced multiple abiotic stress tolerances [23].
At present, the function of WRKY transcription factor has been well elucidated in model plants. However, to some extent, this family has not been well studied in some non-model plants or woody plants. To our knowledge, no information is available about the isolation and functional analysis of WRKY genes in wintersweet. We noted a novel WRKY gene, CpWRKY71, appeared to be differentially expressed during flower development, based on the information from the wintersweet transcriptome database [24]. Here, we isolated and characterized a novel WRKY gene CpWRKY71 from wintersweet. The expression pattern of CpWRKY71 was investigated. Ectopic expression of CpWRKY71 in Arabidopsis resulted in an early flowering and precocious leaf senescence phenotype.
2. Results
2.1. Isolation and Characterization of CpWRKY71
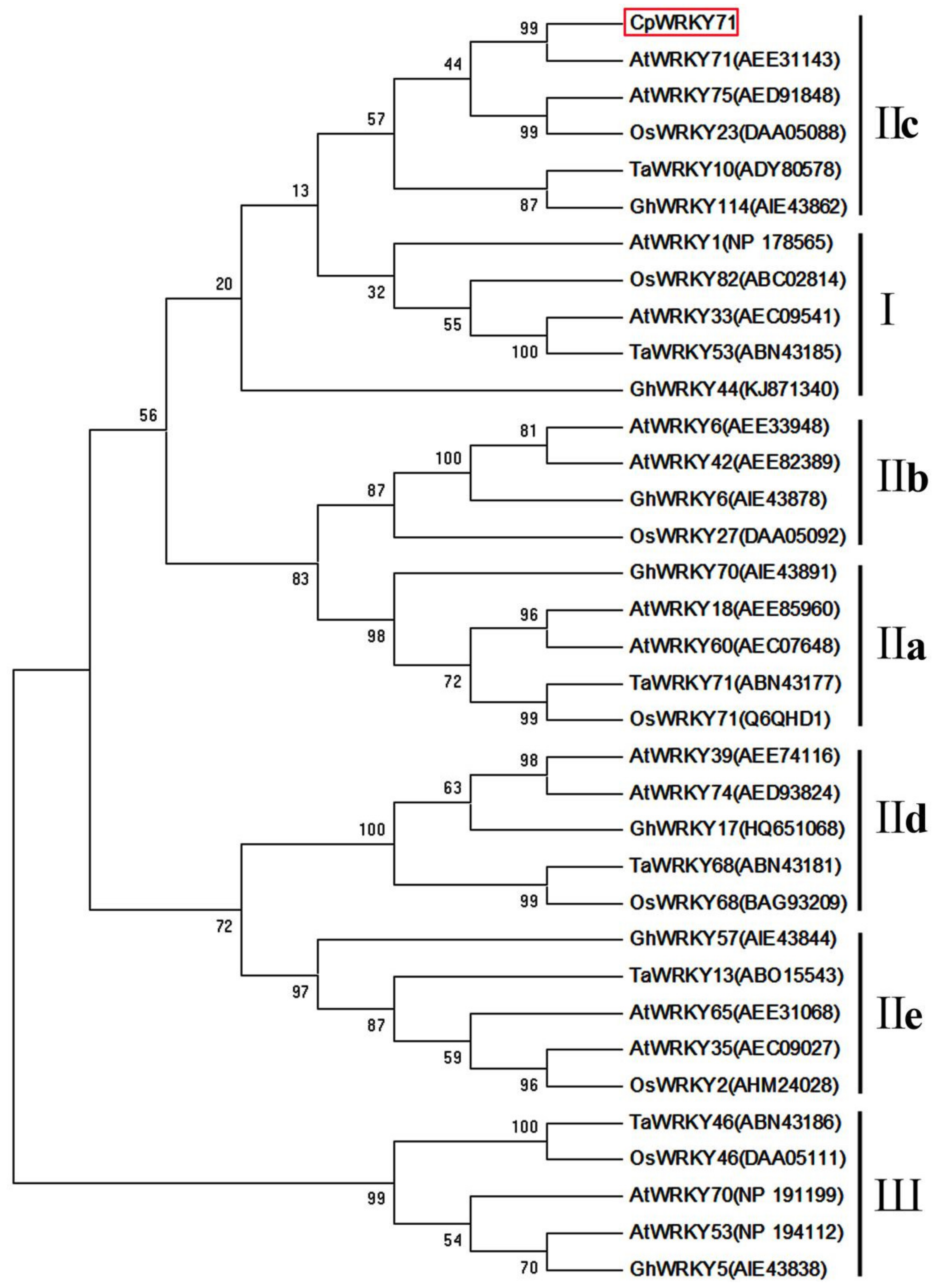
Based on the sequence from the wintersweet flower transcriptome database, a 1137 bp full-length cDNA of CpWRKY71 was obtained. Sequence analysis showed that CpWRKY71 contained a complete open reading frame of 951 bp, which encodes a protein of 316 amino acids. Its predicted molecular weight and theoretical isoelectric point were 34.96 kDa and 6.72, respectively. Subsequently, we isolated 2231 bp genomic DNA fragment of CpWRKY71. Compared with cDNA sequence of CpWRKY71, the genomic DNA of CpWRKY71 contained two introns (417 bp and 677 bp) and three exons (467 bp, 144 bp, and 340 bp) (Figure 1A). Sequence alignments indicated that CpWRKY71 showed high similarity with its homologous sequences, including AtWRKY71(AEE31143), VvWRKY28 (RVW39625), GhWRKY71 (NP_001313893) and NtWRKY71 (XP_009615724). CpWRKY71 contains a single WRKY domain and a C2H2-type zinc finger motif (Figure 1B), indicating that this protein belongs to group II WRKY family. The phylogenetic tree of CpWRKY71 and other WRKY proteins was constructed by MEGA 5.0. The results show that these WRKY proteins can be classified into three groups. CpWRKY71 was clustered with IIc subgroup proteins, including AtWRKY71, AtWRKY75, OsWRKY23, TaWRKY10, GhWRKY114, and was most closely related to AtWRKY71 (Figure 2), so we designated it as CpWRKY71. These results suggest that CpWRKY71 belongs to Group IIc of the WRKY transcription factor family.

Figure 1.
Sequence analysis of CpWRKY71. (A) Schematic diagram of CpWRKY71 gene. Exons are indicated by black areas, introns are indicated by white areas, and untranslated regions are indicated by gray areas. (B) Multiple alignment of CpWRKY71 and other WRKY proteins from different plant species. Arabidopsis thaliana AtWRKY71(AEE31143), Vitis vinifera VvWRKY28 (RVW39625), Gossypium hirsutum GhWRKY71 (NP_001313893), and Nicotiana tomentosiformis NtWRKY71 (XP_009615724) are from GenBank. Identical and similar amino acids were shaded in black and gray, respectively. The conserved WRKY motif and zinc finger motif are marked by the red box.
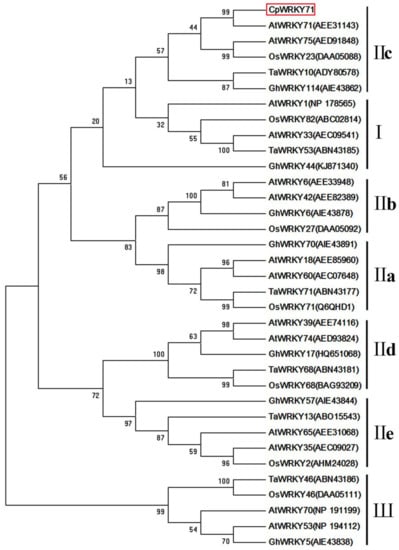
Figure 2.
Phylogenetic tree of CpWRKY71 and WRKY proteins from other plant species. Phylogenetic analysis was performed by the neighbor-joining (NJ) method with 1000 bootstrap replicates using MEGA 5.0. At, Arabidopsis thaliana; Gh, Gossypium hirsutum; Ta, Triticum aestivum; and Os, Oryza sativa. GenBank accession numbers are in the brackets following the protein names. CpWRKY71 is marked by the red box.
2.2. CpWRKY71 is a Nuclear Protein with Transcriptional Activation Activity in Yeast
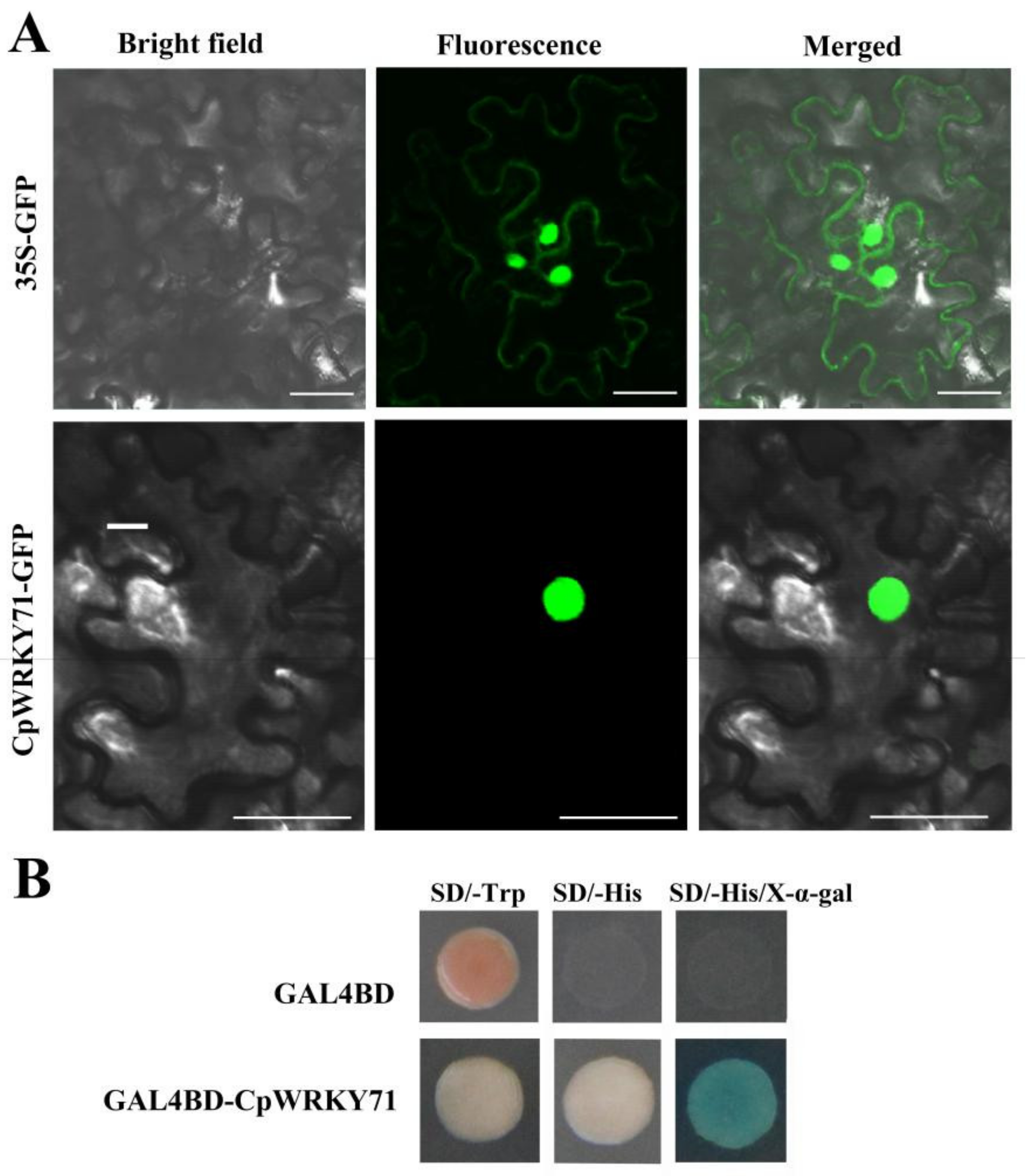
To investigate the subcellular localization of CpWRKY71, CpWRKY71 was fused to the N-terminus of GFP (green fluorescent protein) gene. The Agrobacterium tumefaciens cells harboring 35S:CpWRKY71-GFP or 35S:GFP constructs were infiltrated to young leaves of tobacco (Nicotiana benthamiana) plants, respectively. The GFP fluorescence of CpWRKY71-GFP was observed in the nucleus, whereas that of GFP control was expressed in the cytoplasm and nucleus. These results indicate that CpWRKY71 is a nuclear-localized protein (Figure 3A).
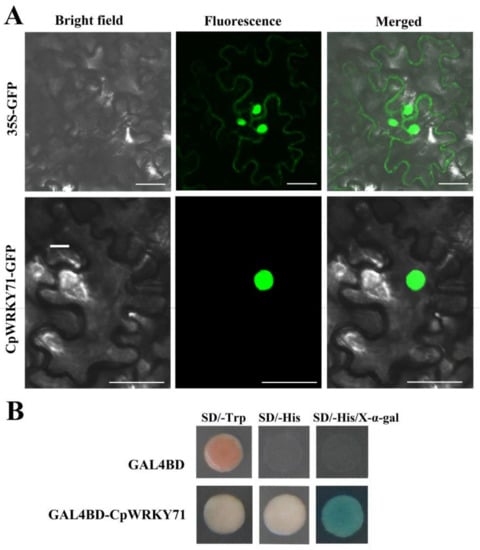
Figure 3.
Subcellular localization and transactivation assay of CpWRKY71. (A) Subcellular localization of CpWRKY71 protein in tobacco leaf epidermal cells. 35S:GFP was used as the control. A confocal microscope was used to observe green fluorescence. Bars denote 50 µm. (B) Transactivation assay of CpWRKY71. The plasmid of pGBKT7-CpWRKY71 was transformed into AH109 yeast strain, and examined on SD/Trp-, SD/-His (containing 10 mM 3-AT) and SD/-His/X-α-gal (containing 10 mM 3-AT) mediums. pGBKT7 was used as the control.
Further, the yeast assay system was used to investigate whether CpWRKY71 has transcriptional activity. The plasmids pGBKT7 and pGBKT7-CpWRKY71 were introduced into AH109 yeast strain, respectively. The results show that yeast cells harboring the plasmid pGBKT7-CpWRKY71 could grow normally on SD/-His medium containing 10 mM 3-amino-1,2,4-triazole (3-AT), and show α-galactosidase activity on the plates containing X-α-gal. On the contrary, the yeast cells transfected with pGBKT7 could survive on the SD/-Trp medium only. This result indicates that CpWRKY71 possesses transcriptional activation activity in yeast (Figure 3B).
2.3. Tissue-Specific Expression of CpWRKY71
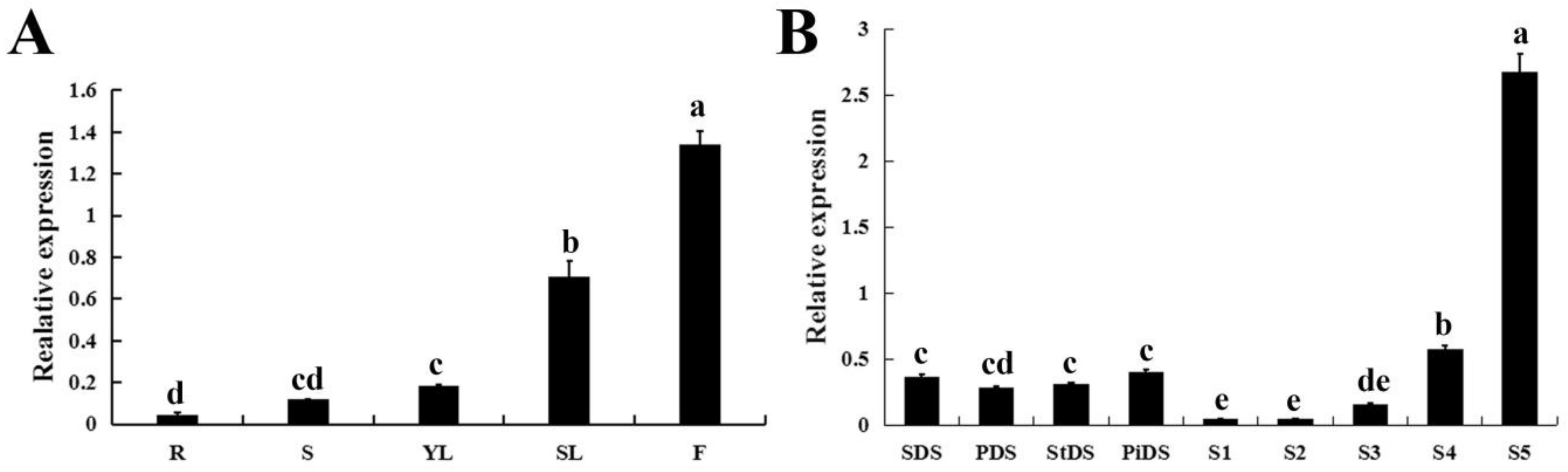
qRT-PCR was performed to analyze the expression pattern of CpWRKY71 in different tissues of wintersweet. The result shows that CpWRKY71 was expressed in all the tissues examined, and the CpWRKY71 transcript was most abundant in flowers. In vegetative organs, the expression level of CpWRKY71 was much higher in senescing leaves than in other tissues (Figure 4A). Moreover, the expression pattern of CpWRKY71 gene during flower development was also examined. The highest CpWRKY71 expression was detected in the early-withering stage (S5), followed by flower primordia differentiation stages (SDS, PDS, StDS, and PiDS) and the bloom stage (S4), and the lowest level was present in the flower-bud (S1), the petal-display (S2) and the initiating bloom stage (S3) (Figure 4B).
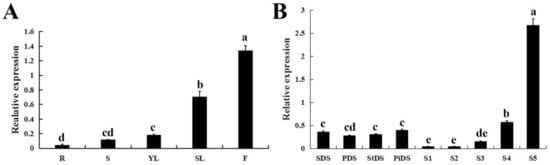
Figure 4.
Expression patterns of CpWRKY71 in wintersweet. (A) The expression pattern of CpWRKY71 in different tissues of wintersweet. R: Roots; S: Stems; YL: Young leaves; SL: Senescing leaves; F: Flowers. (B) The expression pattern of CpWRKY71 in different flower developmental stages of wintersweet. SDS, PDS, StDS and PiDS represent sepal, petal, pistil and stamen primordia the differentiation stage, respectively. S1–S5 represent the flower-bud, the petal-display, the initiating bloom, the bloom, the early-withering stage, respectively. CpActin and CpTublin were used as internal control. Data represent the mean of three biological repeats ± SD. Error bars indicate the standard deviation. Different lowercase letters indicate significant differences (p < 0.05).
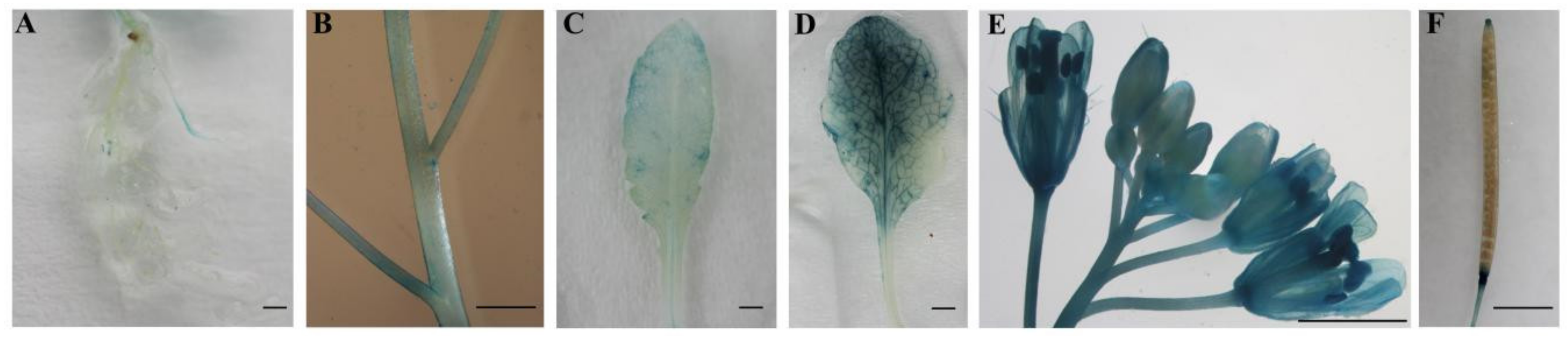
To verify the tissue-specific expression pattern of CpWRKY71 gene, a 3777 bp promoter fragment of CpWRKY71 was fused to the GUS reporter gene to generate transgenic Arabidopsis plants (named CpWRKY71pro:GUS). Histochemical GUS staining showed that faint GUS activity was observed in the roots, stems and young leaves (Figure 5A–C) and strong GUS activity was observed in senescing leaves (Figure 5D). In inflorescence, flower buds showed weak GUS activity. However, GUS staining was increased with the flower blooming, and it was more pronounced in senescing flowers (Figure 5E). In the mature siliques, GUS staining was mainly detected in the abscission zone and the upper portion of the pods (Figure 5F). This result shows that the tissue expression pattern of GUS gene driven by CpWRKY71 promoter in Arabidopsis was similar to that of CpWRKY71 in wintersweet. Collectively, these results show that CpWRKY71 was more highly expressed in senescing flowers and leaves than in other tissues.
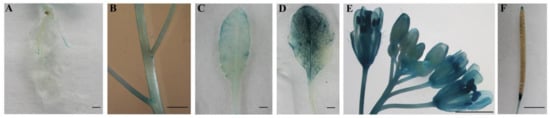
Figure 5.
Tissue-specific GUS activity under the control of the CpWRKY71 promoter. T3 homozygous lines were used for GUS staining analysis. (A) Roots, (B) stems, (C) young leaves, (D) senescing leaves, (E) the whole inflorescence, (F) siliques. Bars denote 2 mm.
2.4. The Expression Profiles of CpWRKY71 under Abiotic Stress and Hormone Treatments
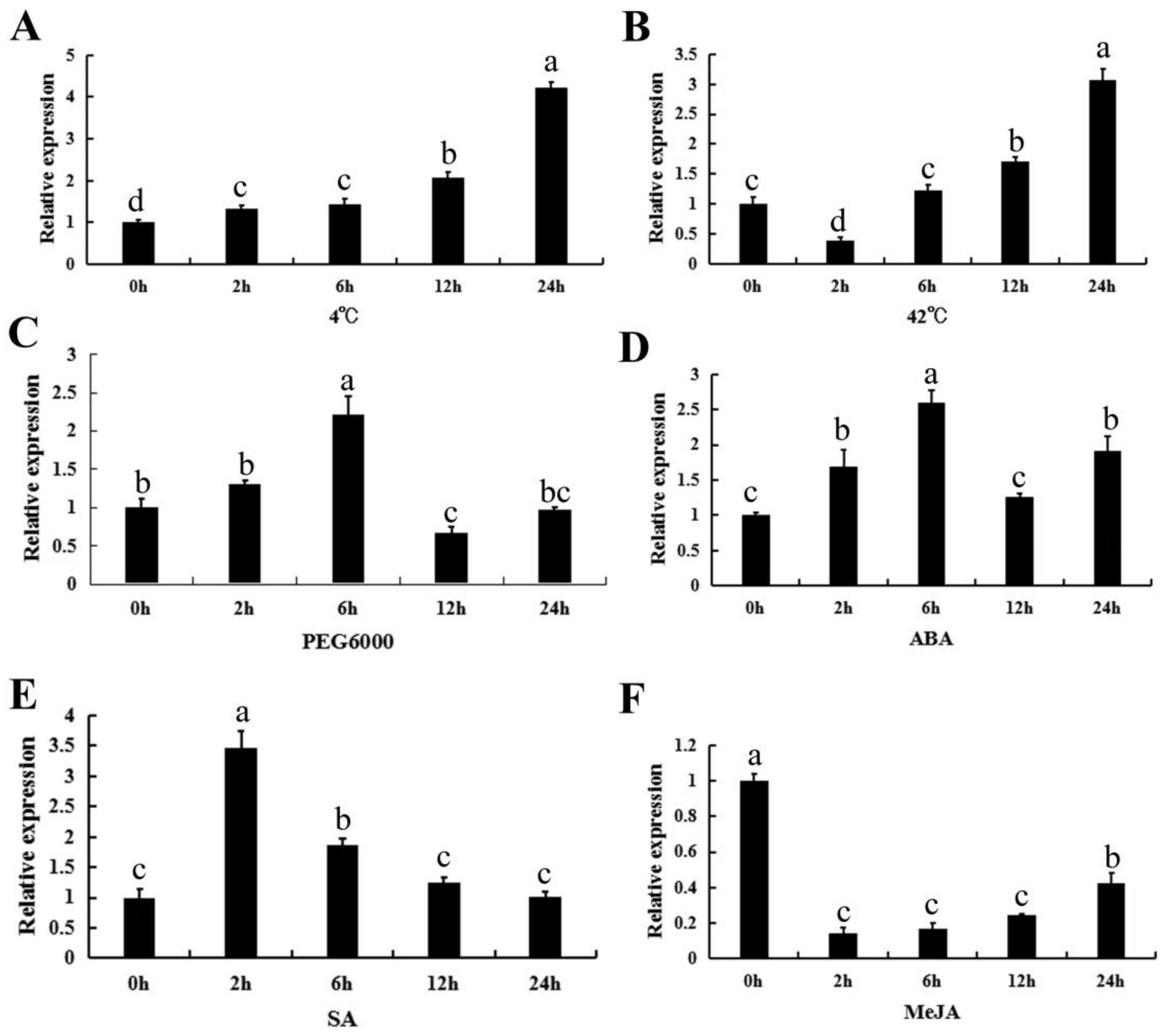
To investigate the effect of stresses and exogenous hormones on CpWRKY71 expression, the transcriptional levels of CpWRKY71 under various abiotic stresses (drought, cold, and heat) and hormone treatments (abscisic acid, salicylic acid, and methyl jasmonate) were detected by qRT-PCR. These treatments were chosen according to the cis-acting elements present in the promoter region of CpWRKY71 (Figure S1). For the cold treatment, the expression of CpWRKY71 was gradually induced and reached a peak at 24 h (Figure 6A). For the heat treatment, CpWRKY71 expression decreased at the 2 h time point, then gradually increased from 6 to 24 h and reached the peak at 24 h (Figure 6B). Under the PEG6000, ABA, and SA treatments, the expression level of CpWRKY71 was induced and peaked at 6, 6, and 2 h respectively, then decreased (Figure 6C–E). For the MeJA treatment, the expression level of CpWRKY71 was decreased dramatically after 2–12 h treatment and then showed a slight increase at 24 h (Figure 6F).
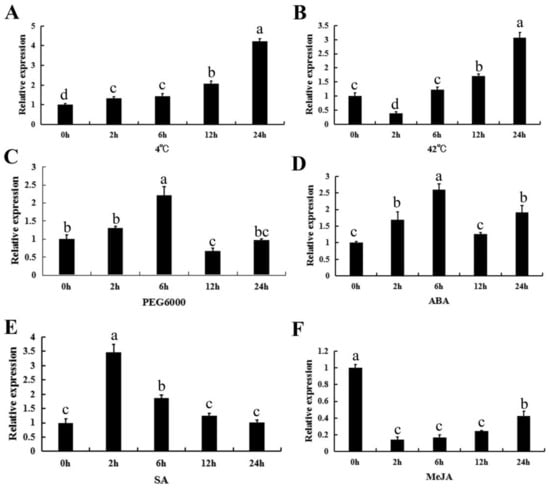
Figure 6.
Expression patterns of CpWRKY71 in response to abiotic stress and hormone treatments. Six-leaf stage wintersweet plants were exposed to treatment with (A) 4 °C, (B) 42 °C, (C) 15% PEG6000, (D) 50 µM ABA, (E) 2 mM SA, and (F) 100 µM MeJA. CpActin and CpTublin were used as internal control. The RNA was extracted from the top leaves at 0, 2, 6, 12 and 24 h post-treatment. Data represent mean of three biological repeats ± SD. Error bars indicate the standard deviation. Different lowercase letters indicate significant differences (p < 0.05).
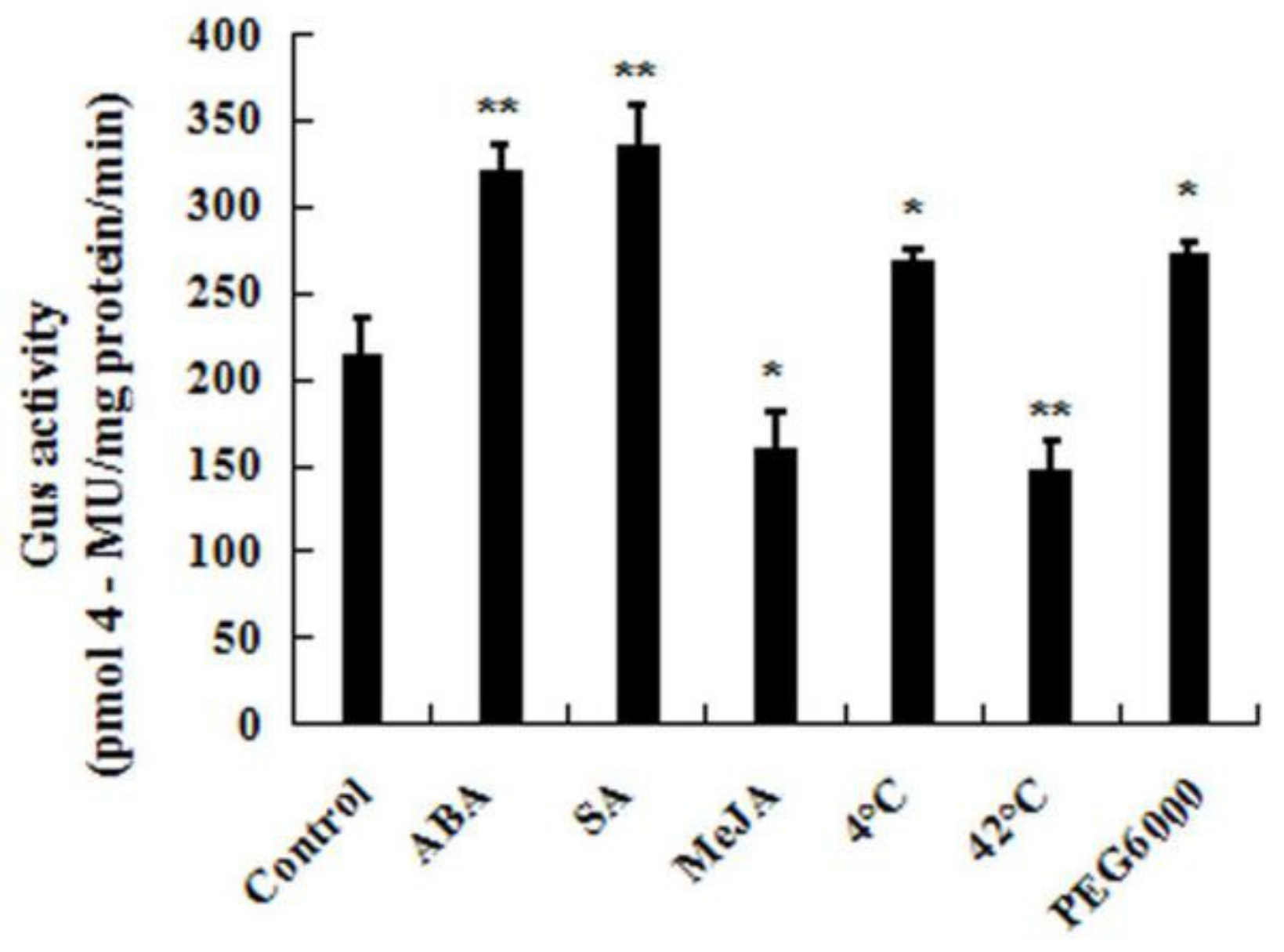
To further study the expression patterns of CpWRKY71 under abiotic stresses and hormone treatments, the responses of CpWRKY71 promoter to the above treatments were also examined (Figure 7). Ten-day-old homozygous CpWRKY71pro:GUS seedlings were used for the assay. The result shows that the GUS activities were significantly increased when seedlings were treated with cold, PEG6000, SA, and ABA. When the plants were treated with heat and MeJA, the GUS enzymatic activity was clearly decreased. Taken with our results, these data suggested that the expression of CpWRKY71 was influenced by various abiotic stresses and hormones.
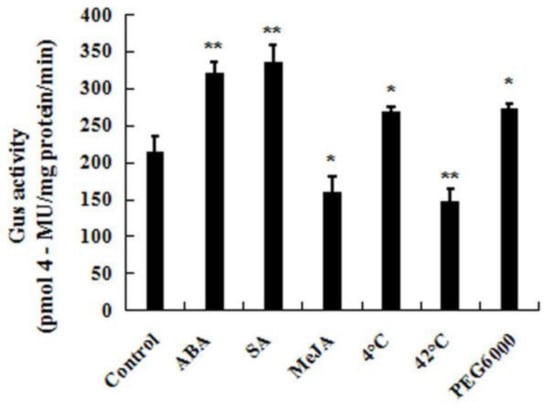
Figure 7.
CpWRKY71 promoter activity in response to abiotic stress and hormone treatments. Ten-day-old T3 homozygous Arabidopsis transgenic seedlings were treated with 50 µM ABA, 100 µM SA, 100 µM MeJA, cold stress at 4 °C and heat stress at 42 °C for 6 h, or 15% PEG6000 for 12 h. The plants grown under normal conditions were used as controls. Data represent the mean of three biological repeats ± SD. Error bars indicate standard deviation. Asterisks denote statistically significant differences compared with control, * p < 0.05, ** p < 0.01.
2.5. Heterologous Overexpression of CpWRKY71 in Arabidopsis Promoted Flowering Time and Leaf Senescence
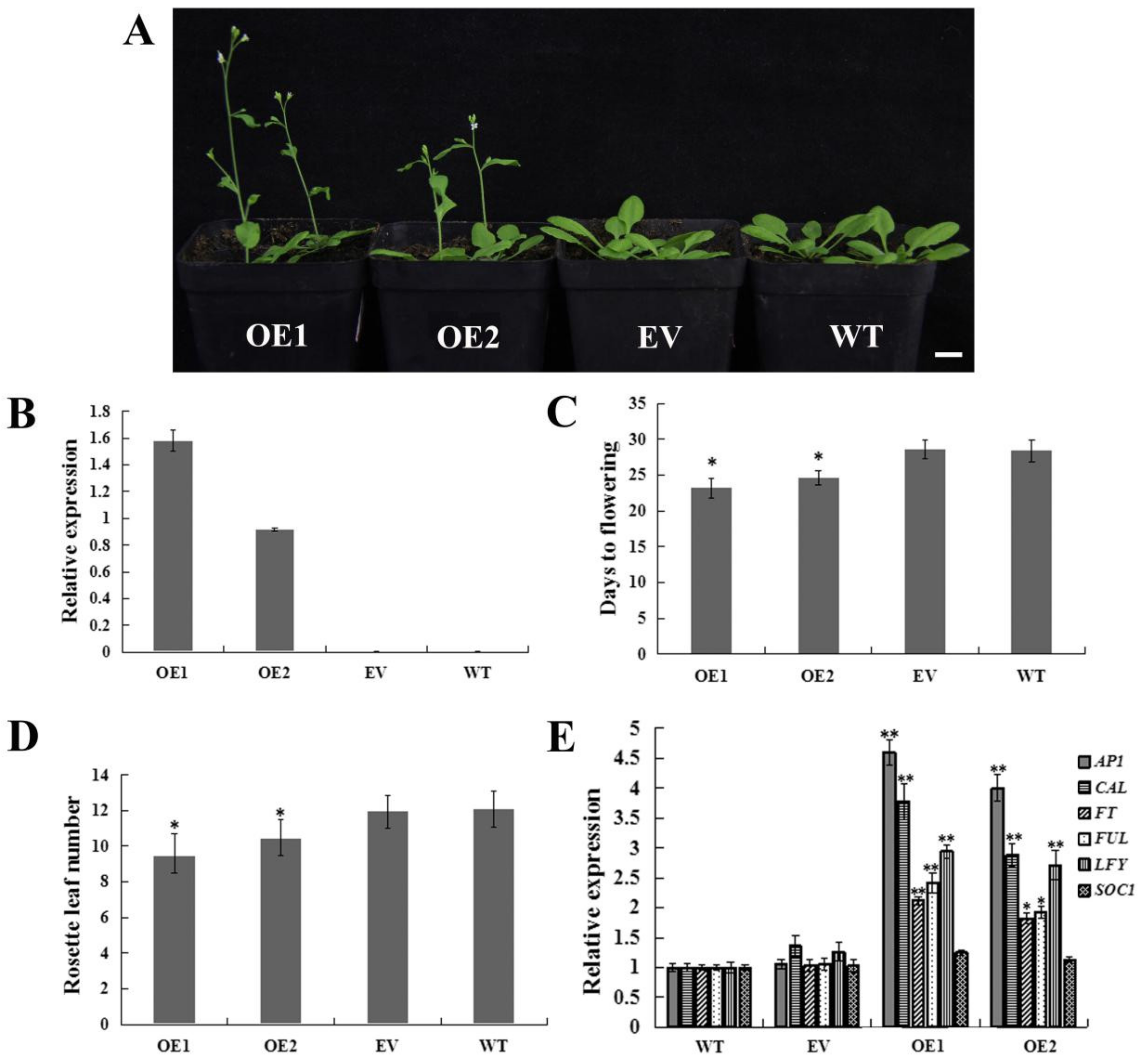
To further investigate the function of CpWRKY71, we transformed the CpWRKY71 gene into Arabidopsis, and dozens of transgenic lines were obtained through hygromycin selection and PCR identification. The expression level was confirmed by qRT-PCR. Two homozygous T3 transgenic lines (OE1 and OE2) were selected for further phenotypic analysis (Figure 8B), the plants transformed with empty vector (EV) and wild type plants (WT) were used as control.
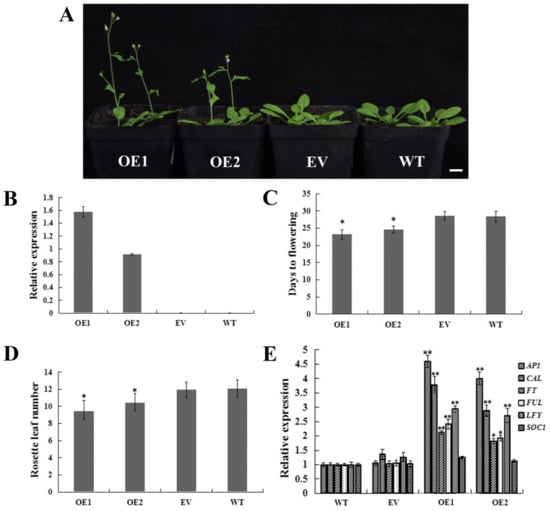
Figure 8.
Flowering phenotype of CpWRKY71 OE plants. (A) The flowering phenotype of 24-day-old plants under LD condition. Bar denotes 1 cm. (B) Transcription levels analysis of CpWRKY71 in different lines. (C) and (D) The flowering time and rosette leaf number of plants grown under LD condition. Data represent the mean ± SD of 20 plants. (E) Expression levels of flowering-related genes in different lines. AtActin was used as an internal control. Data represent mean ± SD of three replicates. Error bars indicate standard deviation. Asterisks denote statistically significant differences compared with control, * p < 0.05, ** p < 0.01.
Under long day (LD) conditions, compared with EV and WT plants, OE1 and OE2 showed an early flowering phenotype (Figure 8A). OE1 and OE2 flowered at 23.2 and 24.6 days after germination (DAG) with 8.6 and 9.93 rosette leaves on average, respectively. While the EV and WT plants flowered at 28.4 and 28.6 DAG with 11.9 and 12.1 rosette leaves on average, respectively (Figure 8C,D). Thus, we examined the transcript levels of floral pathway integrators and the FMI genes FT, LEAFY (LFY), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), AP1, FRUITFULL (FUL) and CAULIFLOWER (CAL) in OE, WT, and EV plants. The result shows that all these genes except SOC1 were upregulated in OE1 and OE2 plants (Figure 8E). These results indicate that overexpression of CpWRKY71 in Arabidopsis promoted flowering.
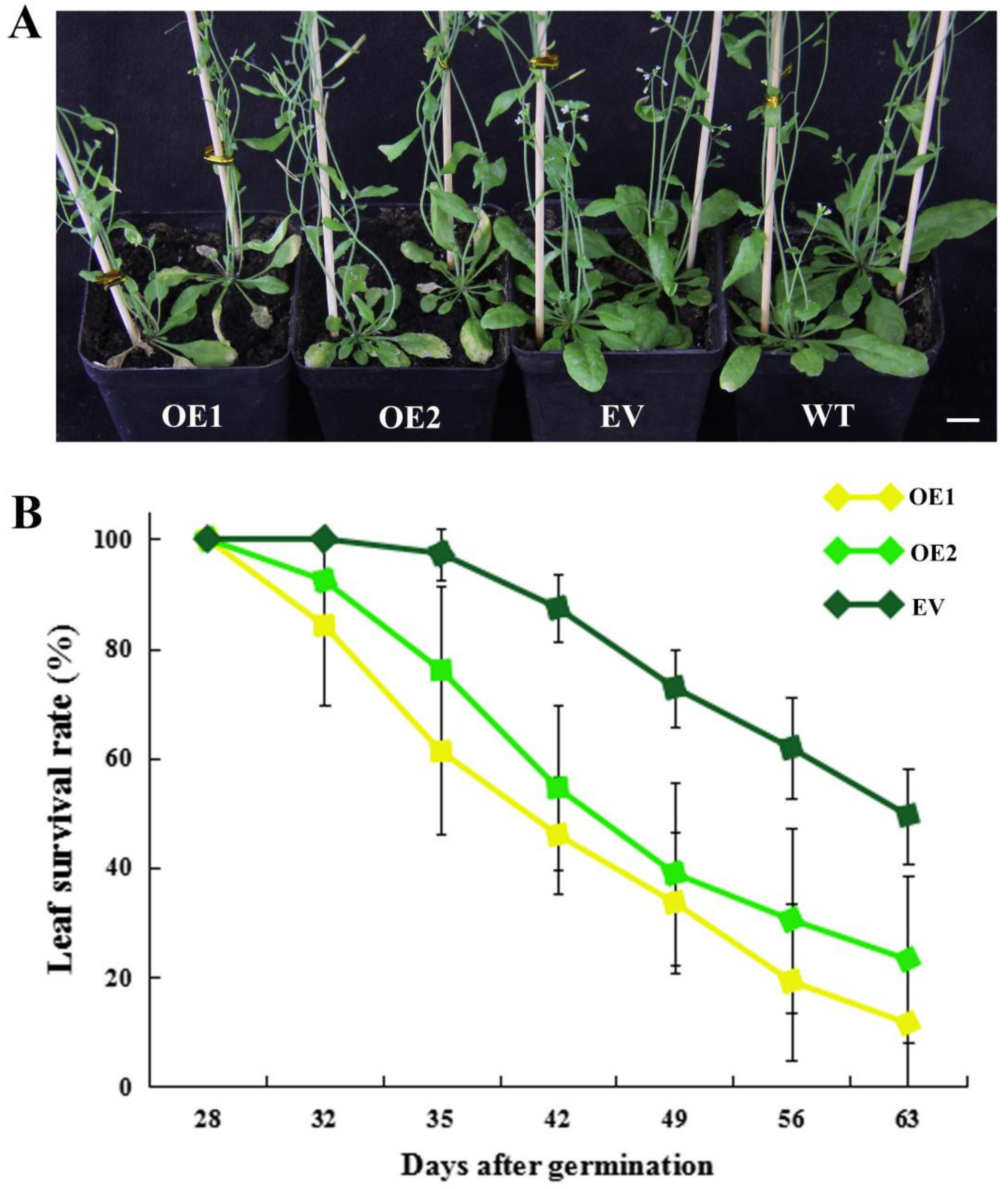
In addition to the early flowering phenotype, we also observed that CpWRKY71 accelerated leaf senescence. OE1 and OE2 exhibited an early leaf senescence phenotype compared with the EV and WT plants (Figure 9A). The leaf survival rates of EV, OE1, and OE2 plants were counted from 28 DAG to 63 DAG. The survival curves further confirmed earlier leaf senescence in transgenic lines than in EV plants (Figure 9B). These data indicate that the constitutive expression of CpWRKY71 accelerated leaf senescence in Arabidopsis.
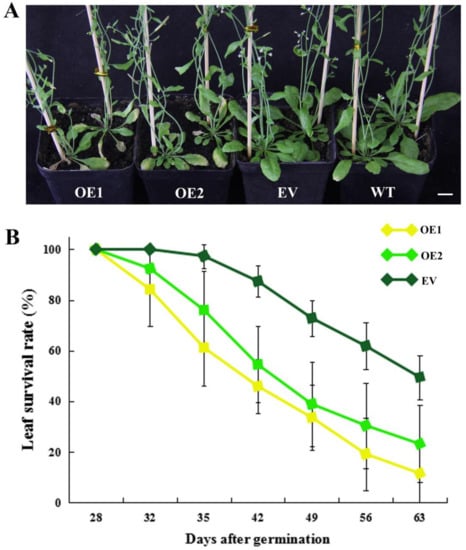
Figure 9.
Precocious leaf senescence phenotype of CpWRKY71 OE plants. (A) The leaf senescence phenotype of 35-day-old plants. Bar denotes 1 cm. (B) Leaf survival curves of OE and EV plants. Data represent the mean ± SD of 20 plants.
3. Discussion
WRKY transcription factors are one of the largest transcription factor families in higher plants. To date, a large number of WRKY transcription factors have been identified in various plant species, such as A. thaliana, Zea mays, Oryza sativa, Glycine max, Populus trichocarpa, and Vitis vinifera [13,14,15,16,17,18]. Wintersweet is a superb winter flowering plant with economic importance in China. However, research about WRKY transcription factors of wintersweet has not yet been reported. In this study, a novel WRKY transcription factor gene, designated CpWRKY71, was isolated from wintersweet and functionally characterized in Arabidopsis. One WRKY domain and a C2H2 zinc finger motif were present in the CpWRKY71 sequence (Figure 1B), which is in accord with the characteristics of group II members. Phylogenetic analysis shows that CpWRKY71 is clustered with group IIc WRKY proteins and has a close relationship with AtWRKY71 (Figure 2). AtWRKY71 was reported to accelerate flowering via the direct activation of FT and LFY in Arabidopsis [25]. Other group members, such as AtWRKY75 was reported to participate in the regulation of flowering in Arabidopsis [26]. In addition, AtWRKY75 was also shown to act as a positive regulator of leaf senescence [27]. This will provide some clues for the functional analysis of CpWRKY71 gene. CpWRKY71 is a nuclear-localized protein and has transcription activation activity in yeast, which was consistent with the results observed in previous reports [25,28], indicating that CpWRKY71 functions as a transcriptional factor (Figure 3).
The expression patterns of a gene can reflect its function to some extent. PtrWRKY19 was highly expressed in stems, especially in the pith, further exploration revealed that PtrWRKY19 participates in the regulation of pith secondary wall formation [29]. The expression of cotton GhWRKY27 was induced by leaf senescence, further functional analysis showed that ectopic expression of GhWRKY27 caused precocious leaf senescence phenotype in transgenic Arabidopsis [30]. In this study, the tissue-specific expression pattern of CpWRKY71 showed that CpWRKY71 expressed higher in flowers and senescing leaves than in other tissues (Figure 4A). During the flower development, high expression of CpWRKY71 was detected in the early-withering stage and obvious expression of CpWRKY71 was also observed in flower primordia differentiation and bloom stages (Figure 4B). These results indicate that CpWRKY71 may play roles in wintersweet flower development and senescence.
Previous reports demonstrated that WRKY genes involved in the response to hormones and abiotic stresses [20,22]. In this study, the expression of CpWRKY71 gene was affected by cold, heat, drought, ABA, SA and MeJA treatments (Figure 6). Corresponding with the expression patterns, abiotic stress and hormone-related cis-acting elements were found in the promoter sequence of CpWRKY71 (Figure S1). Compared with the promoter of CpWRKY71, cis-elements such as ABRE, TCA-element, CGTCA-motif, and TGACG-motif, were also found in the promoter sequence of AtWRKY71. However, different from that of CpWRKY71, cis-regulatory elements such as LTR, HSE and MBS were not observed in the AtWRKY71 promoter (Figure S2). Therefore, the regulatory mechanism of CpWRKY71 in wintersweet might be different from that of AtWRKY71 in Arabidopsis in some respects, even though CpWRKY71 shares some similarity with AtWRKY71.
WRKY proteins have been reported to be involved in various developmental processes, such as trichrome and seed coat development [31], dormancy [19], senescence [20,27], flowering [9,21,25,26], and stress responses [23]. In this study, overexpression of CpWRKY71 in Arabidopsis caused an early flowering phenotype. The expression levels of flowering integrator and FMI genes (FT, LFY, AP1, CAL, and FUL) were increased in transgenic lines (Figure 8). The early flowering phenotype and upregulation of flowering-related genes exhibited some similarities to its orthologue AtWRKY71, this indicates that CpWRKY71 participated in the regulation of flowering time in Arabidopsis. According to several previous studies, different WRKY proteins may be involved in the regulation of flowering in distinct ways. For example, Arabidopsis WRKY12 and WRKY13 were reported to oppositely modulate flowering under SD conditions via directly regulating FUL [32]. AtWRKY71 was found to promote flowering by directly activating the expression of FT and LFY [25]. AtWRKY75 was reported to positively regulate flowering via interaction with DELLA proteins [26]. These results, especially that of AtWRKY71, provide the foundation for further analyzing the role of CpWRKY71 in the control of flowering time. Many WRKY genes have multiple functions in plants [26,27,33,34,35,36]. For example, Miscanthus MlWRKY12 was reported to participate in pith secondary cell wall formation and the regulation of flowering [9]. Rice OsWRKY78 was found to be involved in seed development and stem elongation [37]. In this study, CpWRKY71 overexpression plants showed precocious leaf senescence in addition to the early flowering phenotype (Figure 9), which is in accordance with its expression pattern. Previous studies have shown that some WRKY genes participated in the regulation of leaf senescence. For example, Arabidopsis WRKY54 and WRKY70 were reported to co-operate as negative regulators of leaf senescence [38]. Arabidopsis AtWRKY45 was shown to participate in the regulation of leaf senescence through interaction with DELLA Protein RGL1 [39]. Leaf senescence is influenced by environmental and endogenous cues, such as abiotic stresses and hormones [30]. For instance, Arabidopsis JUB1, an H2O2-induced NAC transcription factor gene, overexpression of JUB1 in Arabidopsis resulted in delayed senescence through lowering the cellular H2O2 level [40]. Arabidopsis S3H which was induced by SA, the S3H gene knockout mutant s3h displayed early leaf senescence phenotype, and S3H regulates leaf longevity by mediating SA catabolism [41]. The foxtail millet SiNAC1 gene expression was induced by ABA and senescence. Functional analysis revealed that SiNAC1 regulates leaf senescence through the ABA pathway [42]. In this study, CpWRKY71 expression was influenced by various stresses and hormones (Figure 6). Therefore, we speculate that CpWRKY71 might affect leaf senescence through regulation of one specific or multiple pathways. In the past decades, many WRKY genes have been reported to affect both flowering and leaf senescence [20,25,26,32]. Taken together, our results indicate that CpWRKY71 may function in regulating flowering time and leaf senescence in transgenic Arabidopsis.
So far, research about WRKY transcription factors of wintersweet has not yet been reported. Hence, our study laid a good foundation for further analysis of WRKY genes in wintersweet. Meanwhile, the functional characterization of CpWRKY71 broadens our knowledge of the roles that WRKY genes may play in woody plants. Moreover, these findings also enrich our knowledge of the molecular mechanism about flowering and senescence in wintersweet.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Seeds of wintersweet used in this research were collected from Southwest University. The seeds were treated with 95–98% sulfuric acid for 30 min. After that, wintersweet seeds were cleaned under running water and then sown into pots filled with a peat and perlite mix (1:1) under greenhouse conditions (16 h light/8 h dark photoperiod, light intensity of 20000 lx, 25 °C).
For the tissue-specific expression pattern of CpWRKY71, the roots, stems, and young leaves were collected from six-leaf stage plants, senescence leaves and flowers at stage 4 (bloom period) were collected from the adult wintersweet plant [1]. Three biological replicates of each tissue were collected.
For the expression pattern of CpWRKY71 gene in different flower development stages, samples at different flower developmental stages were collected as previously described [5]. Three biological replicates of each sample were collected.
Seeds of the A. thaliana ecotype Columbia were sown on solid Murashige and Skoog (MS) medium before vernalization at 4 °C for three days and then grown in a growth chamber at 22 °C with the16 h light/8 h dark photoperiod (120 µmol photons·m−2·s−1).
4.2. Gene Cloning
Total RNA isolation and the first-strand cDNA synthesis were performed as described by Liu et al. [5]. Total genomic DNA was extracted from the wintersweet leaf using CTAB method [43]. Specific primers CpWRKY71-F/R (Table S1) were designed to clone the cDNA and DNA sequence of CpWRKY71. The PCR products were isolated from the gel using the Agarose Gel DNA extraction kit (Tiangen, Beijing, China), then cloned into the pMD19-T vector (TakaRa, Dalian, China) and sequenced by TsingKe Company (TsingKe, Chengdu, China). The multiple alignments were performed using BioEdit software. MEGA 5.0 software with the NJ method (1000 BootStrap replicates) was used to construct the phylogenetic tree of CpWRKY71 and WRKY proteins from other species.
The 5’-upstream sequence of CpWRKY71 was isolated according to the protocol of the Universal Genome Walker Kit (Clontech, USA). Specific primers GSP1 and GSP2 (Table S1) were designed according to the CpWRKY71 gene sequence to clone the promoter. The purification and sequencing of the PCR products were performed as described above. The putative cis-acting regulatory elements were searched with the PLACE database (http://www.dna.affrc.go.jp/PLACE) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [44].
4.3. Plasmid Constructs
For the subcellular localization assay, the open reading frame (ORF) of CpWRKY71 without termination codon was cloned into the modified pCAMBIA 1300 vector [5] via SacI and SalI sites, to construct the plasmid 35S:CpWRKY71-GFP. For transcriptional activity analysis, the ORF of CpWRKY71 was inserted into pGBKT7 via XmaI and Sal I sites, to create the plasmid pGBKT7-CpWRKY71. For GUS histological assay, CpWRKY71 promoter region was cloned into the vector pBI121 to replace the CaMV35S promoter via HindIII and BamHI sites, to construct the plasmid CpWRKY71pro:GUS. The plasmid 35S:CpWRKY71-GFP was also used to generate CpWRKY71 overexpression plants. The primers are listed in Table S1.
4.4. Subcellular Localization and Transactivation Activity Assay of CpWRKY71
The recombinant plasmid 35S:CpWRKY71-GFP and the empty vector 35S:GFP were separately introduced into A. tumefaciens strain GV3101. The young leaves of tobacco (N. benthamiana) plants were infiltrated with A. tumefaciens cells harboring 35S:GFP and 35S:CpWRKY71-GFP, respectively. Confocal laser microscopy (Olympus, Japan) was used to observe the GFP fluorescence.
The recombinant plasmid pGBKT7-CpWRKY71 and pGBKT7 were transformed into the AH109 yeast strain, respectively. SD/-Trp medium was used to select the positive transformants. SD/-His (containing 10 mM 3-AT) and SD/-His/X-α-gal medium (containing 10 mM 3-AT) were used for transactivation analysis. The pGBKT7 empty vector was used as a negative control.
4.5. Plant Treatment
In order to understand gene expression patterns under different abiotic and hormone treatments, wintersweet plants at the six-leaf stage were used [45]. For the drought treatments, plants were irrigated with 15% PEG6000 [46]. For the cold and heat treatments, plants were treated with 4 °C or 42 °C [45], respectively. For exogenous hormone treatments, wintersweet plants were sprayed with 50 µM ABA, 2 mM SA, or 100 µM MeJA [45,47], respectively. Two top leaves of one individual wintersweet plant were collected as one replicate. There were three biological replicates for each treatment. The samples were collected at 0, 2, 6, 12, and 24 h after treatment, frozen in liquid nitrogen and stored at −80 °C ultra- freezer until used.
To understand the effects of different treatments on GUS gene driven by the CpWRKY71 promoter in Arabidopsis, ten-day-old homozygous Arabidopsis T3 seedlings were used [48]. For drought stress, seedlings were transferred into a new plate, then irrigated with 15% PEG6000 for 12 h [49]. For the cold and heat treatments, seedlings were treated with 4 °C or 42 °C for 6 h [48]. For exogenous hormone treatments, Arabidopsis seedlings were sprayed with 50 µM ABA, 100 µM SA, or 100 µM MeJA for 6 h [50,51,52], respectively. The seedlings grown under normal conditions were used as controls. Each experiment contained three biological replicates. These samples were then used for GUS activity analysis.
4.6. Quantitative Real Time-PCR Analysis
qRT-PCR was performed on a Bio-Rad CFX96 Real-time system machine using the Ssofast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA). The PCR procedure was as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 5 s and 72 °C for 5 s, and a melt cycle from 65 to 95 °C. CpActin, CpTubulin, and AtActin were used as the internal reference for the gene expression analysis in wintersweet and Arabidopsis, respectively [5]. The comparative CT method was used to analyze real-time PCR data [5]. All the primers used for qRT-PCR are listed in Table S1.
4.7. Arabidopsis Transformation
For GUS histological assays, the recombinant plasmid CpWRKY71pro:GUS was introduced into A. tumefaciens strain GV3101 and then transformed into Arabidopsis via the floral dipping transformation method [53]. T0 seeds were sowed on MS media containing 50 µg/mL kanamycin. CpWRKY71 overexpression plants were generated as described above. The positive plants were selected on MS media containing 25 µg/mL hygromycin. The seedlings with the resistance grown in a growth chamber under long-day conditions (22 °C with the 16 h light/8 h dark photoperiod and 120 µmol photons·m−2·s−1 light intensity).
4.8. GUS Histochemical and GUS Activity Assays
Histochemical staining was performed according to the method described by Khan et al. [50]. Samples were immersed into GUS staining solution. After incubation in the dark at 37 °C overnight, they were rinsed with 70% (v/v) ethanol to completely remove chlorophyll and then photographed by stereoscopic microscope (Nikon, Japan).
The procedure of the GUS fluorometric assay was performed as described by Niu et al. [49]. The protein concentration of the samples was measured according to the Bradford method (Bradford, 1976) by using Modified Bradford Protein Assay Kit (Sangon, Shanghai, China). 4-Methylumbelliferyl-β-d-glucuronic acid (MUG, Sangon, Shanghai, China) was used as the substrate. The GUS activity was measured using a Varioskan Flash Spectral Scanning Multimode Reader (Thermo Fisher, Waltham, MA, USA) with 365 nm excitation and 455 nm emission. GUS enzyme activity was expressed as pmol 4-MU produced per mg protein per minute.
4.9. Phenotype Analysis
The flowering time was measured as described by Zhang et al. [26]. The day when the first visible flower appeared, and the total rosette leaf number were measured to determine the flowering time. The completely yellowed leaf was regarded as a dead leaf [41].
4.10. Statistical Analysis
Statistical analysis was conducted using SPSS 17.0 software (SPSS, Chicago, IL, USA). The significance of differences was analyzed by Student’s t-test. It was considered significant with a p-value of 0.05, or very significant with a p-value of 0.01.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/21/5325/s1.
Author Contributions
R.H., D.L., J.M., S.S., and M.L. designed the experiments. R.H. and M.H. performed the experiments. R.H., D.L., and J.M. analyzed the data. R.H. wrote the manuscript. D.L., Z.L., S.S., and M.L. edited the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No.31500573), Graduate Scientific Research and Innovation Foundation of Chongqing (No.CYB16070) and Fundamental Research Funds for the Central Universities (XDJK2017B031).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABA | Abscisic acid |
| CTAB | Cetyltrimethylammonium Bromide |
| GUS | β-glucuronidase |
| MeJA | methyl jasmonate |
| PEG6000 | Polyethylene glycol 6000 |
| SA | Salicylic acid |
| qRT-PCR | Quantitative Real Time-PCR |
References
- Ma, J.; Li, Z.; Wang, B.; Sui, S.Z.; Li, M.Y. Cloning of an expansin gene from Chimonanthus praecox flowers and its expression in flowers treated with ethephon or 1-Methylcyclopropene. HortScience 2012, 47, 1472–1477. [Google Scholar] [CrossRef]
- Sui, S.Z.; Luo, J.H.; Ma, J.; Zhu, Q.L.; Lei, X.H.; Li, M.Y. Generation and analysis of expressed sequence tags from Chimonanthus praecox (Wintersweet) flowers for discovering stress-responsive and floral development-related genes. Comp. Funct. Genom. 2012, 2012, 134596. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, B.G.; Duan, K.; Wang, L.G.; Wang, M.; Tang, X.M.; Pan, A.H.; Sui, S.Z.; Wang, G.D. The paleoAP3-type gene CpAP3, an ancestral B-class gene from the basal angiosperm Chimonanthus praecox, can affect stamen and petal development in higher eudicots. Dev. Genes Evol. 2011, 221, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.G.; Duan, K.; Zhang, Q.; Pan, A.H.; Sui, S.Z.; Li, M.Y.; Wang, L.G.; Tang, X.M. The AGL6-like gene CpAGL6, a potential regulator of floral time and organ identity in wintersweet (Chimonanthus praecox). Plant Growth Regul. 2011, 30, 343–352. [Google Scholar] [CrossRef]
- Liu, H.M.; Huang, R.W.; Ma, J.; Sui, S.Z.; Guo, Y.L.; Liu, D.F.; Li, Z.N.; Lin, Y.C.; Li, M.Y. Two C3H type zinc finger protein genes, CpCZF1 and CpCZF2, from Chimonanthus praecox affect Stamen development in Arabidopsis. Genes 2017, 8, 199. [Google Scholar] [CrossRef]
- Luo, J.H.; Ma, J.; Liu, D.F.; Yang, J.F.; Men, W.T.; Wan, C.; Sui, S.Z.; Li, M.Y. Effects of Ethylene on Cut Flower Opening and Senescence and Expression of Ethylene Receptor Genes in Wintersweet (Chimonanthus praecox). Plant Physiol. J. 2015, 51, 253–258. [Google Scholar]
- Sui, S.Z.; Luo, J.H.; Liu, D.F.; Ma, J.; Men, W.T.; Fan, L.; Bai, Y.; Li, M.Y. Effects of Hormone Treatments on Cut Flower Opening and Senescence in Wintersweet (Chimonanthus praecox). Hortscience 2015, 50, 1365–1369. [Google Scholar] [CrossRef]
- Peng, H.; Cheng, H.Y.; Chen, C.; Yu, X.W.; Yang, J.N.; Gao, W.R.; Shi, Q.H.; Zhang, H.; Li, J.G.; Ma, H. A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 2009, 166, 1934–1945. [Google Scholar] [CrossRef]
- Yu, Y.C.; Hu, R.B.; Wang, H.M.; Cao, Y.P.; He, G.; Fu, C.X.; Zhou, G.K. MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci. 2013, 212, 1–9. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Wang, Q.J.; Guo, Z.R. Progresses on plant AP2/ERF transcription factors. Hereditas 2012, 34, 835–847. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 50 upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.F.; Chen, J.; Chen, Y.F.; Wu, L.J.; Xie, D.X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012, 19, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Yin, G.; Xu, H.; Xiao, S.; Qin, Y.; Li, Y.; Yan, Y.; Hu, Y.K. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013, 13, 148. [Google Scholar] [CrossRef]
- He, H.S.; Dong, Q.; Shao, Y.H.; Jiang, H.Y.; Zhu, S.W.; Cheng, B.J.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, W.; Fang, L.C.; Sun, X.M.; Su, L.Y.; Liang, Z.C.; Wang, N.; London, J.P.; Li, S.H.; Xin, H.P. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014, 14, 103. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, X.G.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Gu, L.J.; Li, L.B.; Wei, H.L.; Wang, H.T.; Su, J.J.; Guo, Y.N.; Yu, S.X. Identification of the group IIa WRKY subfamily and the functional analysis of GhWRKY17 in upland cotton (Gossypium hirsutum L.). PLoS ONE 2018, 13, e0191681. [Google Scholar] [CrossRef]
- Cai, Y.H.; Chen, X.J.; Xie, K.; Xing, Q.K.; Wu, Y.W.; Li, J.; Du, C.H.; Sun, Z.X. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS ONE 2014, 9, e102529. [Google Scholar] [CrossRef] [PubMed]
- Li, P.L.; Song, A.P.; Gao, C.Y.; Jiang, J.F.; Chen, S.M.; Fang, W.M.; Zhang, F.; Chen, F.D. The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem. 2015, 95, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.X.; Tian, Y.C.; Liu, X.Z. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.F.; Sui, S.Z.; Ma, J.; Li, Z.N.; Guo, Y.L.; Luo, D.P.; Yang, J.F.; Li, M.Y. Transcriptomic analysis of flower development in wintersweet (Chimonanthus praecox). PLoS ONE 2014, 9, e86976. [Google Scholar] [CrossRef]
- Yu, Y.C.; Liu, Z.H.; Wang, L.; Kim, S.G.; Seo, P.J.; Qiao, M.; Wang, N.; Li, S.; Cao, X.F.; Park, C.M.; et al. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 2016, 85, 96–106. [Google Scholar] [CrossRef]
- Zhang, L.P.; Chen, L.G.; Yu, D.Q. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef]
- Guo, P.K.; Li, Z.H.; Huang, P.X.; Li, B.S.; Shuang, F.; Chu, J.F.; Guo, H.W. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef]
- Kiranmai, K.; Rao, G.L.; Pandurangaiah, M.; Nareshkumar, A.; Reddy, V.A.; Lokesh, U.; Venkatesh, B.; Johnson, A.M.A.; Sudhakar, C. A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 2017, 9, 346. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, X.; Yang, F.; Fan, D.; Jiang, Y.Z.; Luo, K.M. PtrWRKY19, a novel WRKY transcription factor, contributes to the regulation of pith secondary wall formation in Populus trichocarpa. Sci. Rep. 2016, 6, 18643. [Google Scholar] [CrossRef]
- Gu, L.J.; Dou, L.L.; Guo, Y.N.; Wang, H.T.; Li, L.B.; Wang, C.C.; Ma, L.; Wei, H.L.; Yu, S.X. The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2019, 19, 116. [Google Scholar] [CrossRef]
- Johnson, C.S.; Kolevski, B.; Smyth, D.R. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 2002, 14, 1359–1375. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, H.P.; Yu, D.Q. Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under Short-day conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Robatzek, S.; Somssich, I.E. Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 2002, 16, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Rishmawi, L.; Pesch, M.; Juengst, C.; Schauss, A.C.; Schrader, A.; Hulskamp, M. Non-cell-autonomous regulation of root hair patterning genes by WRKY75 in Arabidopsis. Plant Physiol. 2014, 165, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Encinas-Villarejo, S.; Maldonado, A.M.; Amil-Ruiz, F.; de los Santos, B.; Romero, F.; Pliego-Alfaro, F.; Mucloz-Blanco, J.; Caballero, J.L. Evidence for a positive regulatory role of strawberry (Fragaria x ananassa) FaWRKY1 and Arabidopsis AtWRKY75 proteins in resistance. J. Exp. Bot 2009, 60, 3043–3065. [Google Scholar] [CrossRef]
- Zhang, C.Q.; Xu, Y.; Lu, Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 2011, 234, 541–554. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Chen, L.G.; Xiang, S.Y.; Chen, Y.L.; Li, D.B.; Yu, D.Q. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant. 2017, 10, 1174–1189. [Google Scholar] [CrossRef]
- Wu, A.H.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.; Asensi-Fabado, M.A.; Munné-Bosch, S.; Antoni, C.; Tohge, T.; et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Zhang, K.W.; Halitschke, R.; Yin, C.X.; Liu, C.J.; Gan, S.S. Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 2013, 110, 14807–14812. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.T.; Wang, J.W.; Zhao, M.M.; Gong, X.M.; Wang, S.X.; Wang, G.; Zhou, C.J. Involvement of NAC transcription factor SiNAC1 in a positive feedback loop via ABA biosynthesis and leaf senescence in foxtail millet. Planta 2018, 247, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Siegel, C.S.; Stevenson, F.O.; Zimmer, E.A. Evaluation and comparison of FTA card and CTAB DNA extraction methods for non-agricultural taxa. Appl. Plant Sci. 2017, 5, 1600109. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Li, L.; Wei, P.C.; Yang, J.B. Isolation and identifcation of five cold-inducible promoters from Oryza sativa. Planta 2018, 247, 99–111. [Google Scholar] [CrossRef]
- Ma, J.; Men, W.T.; Chen, X.L.; Sui, S.Z.; Li, M.Y. Cloning and expression analysis of a small GTP-binding protein gene (CpRAC1) in Chimonanthus praecox. Acta Hortic. Sin. 2018, 45, 2177–2187. [Google Scholar]
- Yan, H.R.; Jia, H.H.; Chen, X.B.; Hao, L.L.; An, H.L.; Guo, X.Q. The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef]
- Li, J.; Wang, J.; Wang, N.X.; Guo, X.Q.; Gao, Z. GhWRKY44, a WRKY transcription factor of cotton, mediates defense responses to pathogen infection in transgenic Nicotiana benthamiana. Plant Cell Tissue Org. 2015, 121, 127–140. [Google Scholar] [CrossRef]
- Wang, F.Q.; Yang, J.; Dai, C.; Wu, M.Z.; Zhang, Y.H.; Shen, W.B. Characterization of the Arabidopsis thaliana heme oxygenase 1 promoter in response to salinity, iron deficiency, and mercury exposure. Biol. Plant. 2017, 61, 35–47. [Google Scholar] [CrossRef]
- Niu, G.L.; Gou, W.; Han, X.L.; Qin, C.; Zhang, L.X.; Abomohra, A.E.; Ashraf, M. Cloning and functional analysis of phosphoethanolamine methyltransferase promoter from Maize (Zea mays L.). Int. J. Mol. Sci. 2018, 19, 191. [Google Scholar] [CrossRef]
- Khan, M.A.; Meng, Y.L.; Liu, D.F.; Tang, H.S.; Lü, S.H.; Imtiaz, M.; Jiang, G.M.; Lü, P.T.; Ji, Y.Q.; Gao, J.P.; et al. Responses of rose RhACS1 and RhACS2 promoters to abiotic stresses in transgenic Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 795–804. [Google Scholar] [CrossRef]
- Ma, J.; Li, Z.; Chen, X.L.; Zhang, S.Y.; Sui, S.Z.; Li, M.Y. Cloning and activity analysis of CpEXP1 gene promoter from Chimonanthus praecox. Sci. Silvae Sin. 2018, 54, 61–72. [Google Scholar]
- Tao, Y.; Wang, F.T.; Jia, D.M.; Li, J.T.; Zhang, Y.M.; Jia, C.G.; Wang, D.P.; Pan, H.Y. Cloning and Functional Analysis of the Promoter of a Stress-inducible Gene (ZmRXO1) in Maize. Plant Mol. Biol. Rep. 2015, 33, 200–208. [Google Scholar] [CrossRef]
- Clough, S.J.; Ben, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).