Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in Solanum tuberosum
Abstract
1. Introduction
2. Results
2.1. Identification of Potato LBD Gene Family Members
2.2. Chromosome Localization of the Potato LBD Gene Family
2.3. Phylogenetic Evolution and Gene Structure Analysis of the Potato LBD Gene Family
2.4. Conservative Analysis of the Potato LBD Protein Sequence
2.5. Tissue Expression and Induced Expression Analysis of the Potato LBD Gene
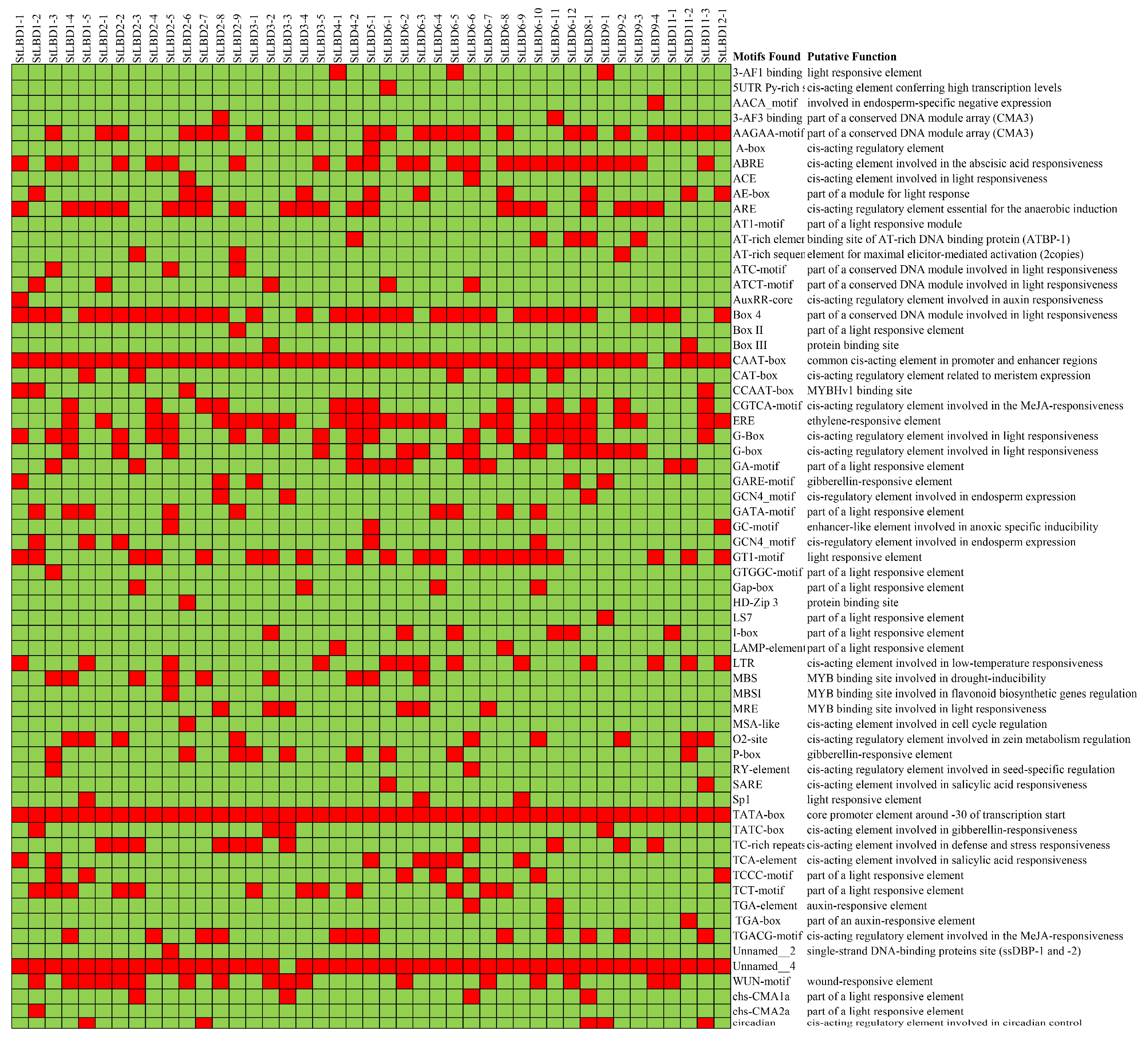
2.6. Analysis of Cis-Acting Elements of the Potato LBD Gene
2.7. Expression Analysis of Seven LBD Genes in the Leaf at Different Stages
2.8. Expression Analysis of Seven LBD Genes in Different Tissues
3. Discussion
4. Materials and Methods
4.1. Identification and Chromosomal Localization of Potato LBD Family Members
4.2. Phylogenetic Tree Construction, Gene Structure Analysis and Protein Domain Sequence Alignment
4.3. Promoter Cis-Acting Element Analysis
4.4. Tissue Expression and Induced Expression Analysis of the Potato LBD Gene
4.5. Plant Material, Growth Conditions and Stress Treatment
4.6. Total RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Matsumura, Y.; Iwakawa, H.; Machida, Y.; Machida, C. Characterization of genes in the Asymmetric Leaves2/Lateral Organ Boundaries (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J. 2009, 58, 525–537. [Google Scholar] [CrossRef]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zhao, J. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 2010, 61, 2647–2668. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Liu, X.; Su, L.; Sun, Y.J.; Zhang, S.Z.; Hao, Y.J.; You, C.H. Identification, evolution and expression analysis of the LBD gene family in tomato. Sci. Agric. Sin. 2013, 46, 2501–2513. [Google Scholar]
- Wang, X.; Zhang, S.; Su, L.; Liu, X.; Hao, Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE 2013, 8, e57044. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Liu, W.; Xie, W.; Liu, Z.; Liu, Z.; Wang, Y. Genome-wide analysis of the LBD transcription factor family in Medicago truncatula. Acta Bot. Boreali-Occident. Sin. 2014, 34, 2176–2187. [Google Scholar]
- Zhang, Y.M.; Zhang, S.Z.; Zheng, C.C. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet. 2014, 93, 79–91. [Google Scholar] [CrossRef]
- Luo, Y.; Ma, B.; Zeng, Q.; Xiang, Z.; He, N. Identification and characterization of Lateral Organ Boundaries Domain genes in mulberry, Morus notabilis. Meta Gene 2016, 8, 44–50. [Google Scholar] [CrossRef]
- Zheng, Z.F.; Zhang, Y.L.; Liu, F.; Yuan, Z.H. Genome-wide Identification and Expressing Analysis of LBD Transcription Factors in Pepper. Acta Hortic. Sin. 2016, 43, 683–694. [Google Scholar]
- Cao, H.U.; Liu, C.Y.; Liu, C.X.; Zhao, Y.L.; Xu, R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016, 95, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Liu Tongjin, Z.X.Z.X. Identification and Analysis of LBD Gene Family Members in Radish Whole Genome. J. Plant Genet. Resour. 2019, 1, 168–178. [Google Scholar]
- Lu, Q.; Shao, F.; Macmillan, C.; Wilson, I.W.; Van der Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef]
- Guo, B.J.; Wang, J.; Lin, S.; Tian, Z.; Zhou, K.; Luan, H.Y.; Lyu, C.; Zhang, X.Z.; Xu, R.G. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ. Sci. B 2016, 17, 763–774. [Google Scholar] [CrossRef]
- Chen, W.F.; Wei, X.B.; Rety, S.; Huang, L.Y.; Liu, N.N.; Dou, S.X.; Xi, X.G. Structural analysis reveals a “molecular calipers mechanism” for a Lateral Organ Boundaries Domain transcription factor protein from wheat. J. Biol. Chem. 2019, 294, 142–156. [Google Scholar] [CrossRef]
- Meng, L.S.; Liu, H.L.; Cui, X.; Sun, X.D.; Zhu, J. Asymmetric Leaves2-Like 38 gene, a Member of AS2/LOB family of Arabidopsis, causes leaf dorsoventral alternation in transgenic cockscomb plants. Acta Physiol. Plant. 2009, 31, 1301. [Google Scholar] [CrossRef]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The asymmetric leaves 2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar]
- Iwakawa, H.; Iwasaki, M.; Kojima, S.; Ueno, Y.; Soma, T.; Tanaka, H.; Semiarti, E.; Machida, Y.; Machida, C. Expression of the asymmetric leaves 2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 2007, 51, 173–184. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, M.; Kim, J. Combinatorial interactions between LBD10 and LBD27 are essential for male gametophyte development in Arabidopsis. Plant Signal. Behav. 2015, 10, e1044193. [Google Scholar] [CrossRef]
- Luo, J.; Weng, L.; Luo, D. Isolation and Expression Patterns of LATERAL ORGAN BOUNDARIES-like Genes in Lotus japonicus. J. Plant Physiol. Mol. Biol. 2006, 32, 202. [Google Scholar]
- Zhang, J.; Tang, W.; Huang, Y.; Niu, X.; Zhao, Y.; Han, Y.; Liu, Y. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J. Exp. Bot. 2015, 66, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H.; Liu, X.; An, J.P.; Hao, Y.J.; Wang, X.F.; You, C.X. Cloning and elucidation of the functional role of apple MdLBD13 in anthocyanin biosynthesis and nitrate assimilation. Plant Cell Tissue Organ Cult. 2017, 130, 47–59. [Google Scholar] [CrossRef]
- Evans, M.M.S. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 2007, 19, 46–62. [Google Scholar] [CrossRef]
- He, H.H.; Ma, Z.; Zhang, Y.; Zhang, J.; Lu, S.; Zhang, Z.; Zhao, X.; Wu, Y.; Mao, J. Identification and Expression Analysis of LBD Gene Family in Grape. Sci. Agric. Sin. 2018, 51, 4102–4118. [Google Scholar]
- Zebarth, B.J.; Tai, H.; Luo, S.; Millard, P.; De Koeyer, D.; Li, X.Q.; Xiong, X. Effect of Nitrogen Form on Gene Expression in Leaf Tissue of Greenhouse Grown Potatoes During Three Stages of Growth. Am. J. Potato Res. 2012, 89, 315–327. [Google Scholar] [CrossRef]
- Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Bdeir, R.; Busov, V.; Yordanov, Y.; Gailing, O. Gene dosage effects and signatures of purifying selection in lateral organ boundaries domain (LBD) genes LBD1 and LBD18. Plant Syst. Evol. 2016, 302, 433–445. [Google Scholar] [CrossRef]
- Messing, J.; Geraghty, D.; Heidecker, G.; Hu, N.T.; Kridl, J.; Rubenstein, I. Plant Gene Structure. In Genetic Engineering of Plants; Springer: Boston, MA, USA, 1983. [Google Scholar]
- Nevins, J.R. The pathway of eukaryotic mRNA formation. Annu. Rev. Biochem. 1983, 52, 441–466. [Google Scholar] [CrossRef]
- Lam, E.; Chua, N.H. ASF-2: A factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1989, 1, 1147–1156. [Google Scholar] [PubMed]
- Terzaghi, W.B.; Cashmore, A.R. Light-Regulated Transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol 1995, 46, 445–474. [Google Scholar] [CrossRef]
- Liu, L.; Xu, W.; Hu, X.; Liu, H.; Lin, Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016, 6, 20881. [Google Scholar] [CrossRef] [PubMed]
- Heidari, P.; Ahmadizadeh, M.; Najafi-Zarrini, H. In Silico Analysis of Cis-Regulatory Elements on Co-Expressed Genes. J. Biol. Env. Sci. 2015, 9, 1–9. [Google Scholar]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14 (Suppl. S1), S15–S45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Zhang, H.; Li, R.; Hong, Q.; Li, Y.; Xia, Q.; Zhang, W. Function Identification of the Nucleotides in Key cis-Element of DYSFUNCTIONAL TAPETUM1 (DYT1) Promoter. Front Plant Sci. 2017, 8, 153. [Google Scholar] [CrossRef]
- Liu, B.; Zhao, S.; Wu, X.; Wang, X.; Nan, Y.; Wang, D.; Chen, Q. Identification and Characterization of phosphate transporter genes in potato. J. Biotechnol. 2017, 264, 17–28. [Google Scholar] [CrossRef]
- Gao, J.; Cao, X.; Shi, S.; Ma, Y.; Wang, K.; Liu, S.; Chen, D.; Chen, Q.; Ma, H. Genome-wide survey of Aux/IAA gene family members in potato (Solanum tuberosum): Identification, expression analysis, and evaluation of their roles in tuber development. Biochem. Biophys. Res. Commun. 2016, 471, 320–327. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, D.; Wang, R.; Kong, N.; Zhang, C.; Yang, C.; Wu, W.; Ma, H.; Chen, Q. Genome-wide analysis of the potato Hsp20 gene family: Identification, genomic organization and expression profiles in response to heat stress. BMC Genom. 2018, 19, 61. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, P.; Kong, N.; Lu, R.; Pei, Y.; Huang, C.; Ma, H.; Chen, Q. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes 2018, 9, 54. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H.; et al. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef] [PubMed]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Zhang, G.; Wu, M.; Wang, G. Identification and characterization of the Populus trichocarpa CLE family. BMC Genom. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Fenjuan, S.; Deyou, Q. Genome-Wide Analysis of Gene Family of Lateral Organ Boundaries Domain in Populus trichocarpa. Genom. Appl. Biol. 2018, 37, 313–315. [Google Scholar]
- Sun, T.T.; Gong, D.P.; Zhang, L.; Chen, Y.Q.; Zhao, W.; Xiang, X.H.; Sun, Y. Genome-wide Sequence Identification and Expression Analysis of the LBD Gene Family in Nicotiana tabacum. J. Plant Genet. Resour. 2016, 17, 316–325. [Google Scholar]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071. [Google Scholar] [CrossRef]
- Chalfun-Junior, A.; Franken, J.; Mes, J.J.; Marsch-Martinez, N.; Pereira, A.; Angenent, G.C. Asymmetric leaves2-like1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Mol. Biol. 2005, 57, 559–575. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, F.; Guo, J.; Zhang, X.S. Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J. Plant Biol. 2009, 52, 374–381. [Google Scholar] [CrossRef]
- Cho, C.; Jeon, E.; Pandey, S.K.; Ha, S.H.; Kim, J. LBD13 positively regulates lateral root formation in Arabidopsis. Planta 2019, 249, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Cavalcanti, A.; Chen, F.C.; Bouman, P.; Li, W.H. Extent of Gene Duplication in the Genomes of Drosophila, Nematode, and Yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef] [PubMed]










| Gene Features Phytozome Gene ID | Phytozome Transcript ID | Protein Features | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Families | Putative Gene Name | Exon No. | Chr. Location | Protein Length | MW (kDa) | pI | GRAVY | TMD/Terminus | ||
| PGSC0003DMG400014757 | PGSC0003DMT400038248* | Ic | StLBD1-1 | 2 | chr01 | 128 | 15 | 5.53 | −0.293 | 0/out→out |
| PGSC0003DMG400012757 | PGSC0003DMT400033221* | Ib | StLBD1-2 | 2 | chr01 | 133 | 15 | 6 | 0.187 | 0/out→out |
| PGSC0003DMG402012772 | PGSC0003DMT400033253* | Ib | StLBD1-3 | 1 | chr01 | 139 | 14 | 9.12 | 0.136 | 0/out→out |
| PGSC0003DMG400022454 | PGSC0003DMT400057829* | Ib | StLBD1-4 | 2 | chr01 | 241 | 26 | 6.13 | −0.393 | 0/out→out |
| PGSC0003DMG400025752 | PGSC0003DMT400066165* | IIa | StLBD1-5 | 2 | chr01 | 217 | 24 | 6.9 | −0.267 | 0/out→out |
| PGSC0003DMG401000764 | PGSC0003DMT400002011* | Ia | StLBD2-1 | 2 | chr02 | 288 | 33 | 6.44 | −0.839 | 0/out→out |
| PGSC0003DMG402000764 | PGSC0003DMT400002013* | Ia | StLBD2-2 | 2 | chr02 | 287 | 33 | 6.23 | −0.806 | 0/out→out |
| PGSC0003DMG400006914 | PGSC0003DMT400017806* | Ia | StLBD2-3 | 1 | chr02 | 168 | 19 | 8.89 | −0.649 | 0/out→out |
| PGSC0003DMG400021204 | PGSC0003DMT400054637* | Ic | StLBD2-4 | 2 | chr02 | 161 | 18 | 8.21 | −0.442 | 0/out→out |
| PGSC0003DMG400012653 | PGSC0003DMT400032938* | IIa | StLBD2-5 | 2 | chr02 | 273 | 30 | 7.02 | −0.392 | 0/out→out |
| PGSC0003DMG400024936 | PGSC0003DMT400064185* | IIa | StLBD2-6a | 2 | chr02 | 212 | 23 | 6.11 | −0.231 | 0/out→out |
| PGSC0003DMT400064186 | IIa | StLBD2-6b | 2 | chr02 | ||||||
| PGSC0003DMG400010023 | PGSC0003DMT400025959* | Ia | StLBD2-7 | 1 | chr02 | 313 | 35 | 5.98 | −0.641 | 0/out→out |
| PGSC0003DMG400038771 | PGSC0003DMT400089200* | Ie | StLBD2-8 | 1 | chr02 | 123 | 15 | 8.93 | −0.658 | 0/out→out |
| PGSC0003DMG400012629 | PGSC0003DMT400032877* | Ia | StLBD2-9 | 1 | chr02 | 157 | 18 | 5.91 | −0.719 | 0/out→out |
| PGSC0003DMG400000974 | PGSC0003DMT400002549* | Ia | StLBD3-1 | 1 | chr03 | 189 | 21 | 8.77 | −0.529 | 0/out→out |
| PGSC0003DMG400027718 | PGSC0003DMT400071281* | Ic | StLBD3-2 | 1 | chr03 | 177 | 20 | 6.29 | −0.388 | 0/out→out |
| PGSC0003DMG400018112 | PGSC0003DMT400046644* | Ia | StLBD3-3 | 1 | chr03 | 191 | 21 | 8.21 | −0.469 | 0/out→out |
| PGSC0003DMG400013182 | PGSC0003DMT400034288* | Ie | StLBD3-4 | 2 | chr03 | 301 | 34 | 5.48 | −0.579 | 0/out→out |
| PGSC0003DMG400005719 | PGSC0003DMT400014630* | IIa | StLBD3-5 | 2 | chr03 | 244 | 27 | 6.44 | −0.361 | 0/out→out |
| PGSC0003DMG400004138 | PGSC0003DMT400010597* | Ic | StLBD4-1 | 2 | chr04 | 173 | 20 | 5.71 | −0.467 | 0/out→out |
| PGSC0003DMG400021509 | PGSC0003DMT400055393* | IIa | StLBD4-2a | 2 | chr04 | 204 | 22 | 5.07 | −0.208 | 0/out→out |
| PGSC0003DMT400055392 | IIa | StLBD4-2b | 2 | chr04 | ||||||
| PGSC0003DMG400014547 | PGSC0003DMT400037711* | IIa | StLBD5-1 | 2 | chr05 | 253 | 28 | 5.47 | −0.426 | 0/out→out |
| PGSC0003DMG400020562 | PGSC0003DMT400052995* | Ic | StLBD6-1 | 2 | chr06 | 201 | 22 | 6.81 | −0.209 | 0/out→out |
| PGSC0003DMG400025400 | PGSC0003DMT400065346* | Ic | StLBD6-2a | 2 | chr06 | 209 | 23 | 5.16 | −0.273 | 0/out→out |
| PGSC0003DMT400065345 | Ic | StLBD6-2b | 1 | chr06 | ||||||
| PGSC0003DMG400046838 | PGSC0003DMT400097267* | Ib | StLBD6-3 | 2 | chr06 | 155 | 18 | 5.4 | −0.051 | 0/out→out |
| PGSC0003DMG400023939 | PGSC0003DMT400061511* | Ic | StLBD6-4 | 2 | chr06 | 171 | 19 | 5.73 | −0.18 | 0/out→out |
| PGSC0003DMG400029080 | PGSC0003DMT400074783* | Ic | StLBD6-5a | 3 | chr06 | 274 | 30 | 4.98 | −0.469 | 0/out→out |
| PGSC0003DMT400074784 | Ic | StLBD6-5b | 2 | chr06 | ||||||
| PGSC0003DMG400004880 | PGSC0003DMT400012498* | Ie | StLBD6-6 | 2 | chr06 | 327 | 37 | 5.48 | −0.895 | 0/out→out |
| PGSC0003DMG400027078 | PGSC0003DMT400069646* | Ia | StLBD6-7 | 1 | chr06 | 175 | 19 | 6.29 | −0.485 | 0/out→out |
| PGSC0003DMG400007503 | PGSC0003DMT400019410* | Ic | StLBD6-8 | 2 | chr06 | 198 | 22 | 6.71 | −0.282 | 0/out→out |
| PGSC0003DMG400030456 | PGSC0003DMT400078256* | Ic | StLBD6-9 | 2 | chr06 | 173 | 19 | 6.7 | −0.221 | 0/out→out |
| PGSC0003DMG400030461 | PGSC0003DMT400078265* | Ic | StLBD6-10a | 2 | chr06 | 222 | 24 | 9.02 | −0.209 | 0/out→out |
| PGSC0003DMT400078264 | Ic | StLBD6-10b | 2 | chr06 | ||||||
| PGSC0003DMG400027663 | PGSC0003DMT400071132* | Ic | StLBD6-11 | 1 | chr06 | 134 | 15 | 4.87 | −0.35 | 0/out→out |
| PGSC0003DMG400020069 | PGSC0003DMT400051680* | Ib | StLBD6-12 | 2 | chr06 | 195 | 22 | 6.49 | −0.341 | 0/out→out |
| PGSC0003DMG400044009 | PGSC0003DMT400094438* | Ie | StLBD8-1 | 1 | chr08 | 302 | 33 | 8.85 | −0.441 | 0/out→out |
| PGSC0003DMG400008500 | PGSC0003DMT400021914* | IIa | StLBD9-1 | 3 | chr09 | 108 | 12 | 5.39 | −0.81 | 0/out→out |
| PGSC0003DMG400043103 | PGSC0003DMT400093532* | IIa | StLBD9-2 | 2 | chr09 | 167 | 19 | 4.9 | −0.559 | 0/out→out |
| PGSC0003DMG400009713 | PGSC0003DMT400025135* | Ib | StLBD9-3 | 2 | chr09 | 221 | 25 | 8.68 | −0.484 | 0/out→out |
| PGSC0003DMG400009716 | PGSC0003DMT400025142* | Ib | StLBD9-4 | 2 | chr09 | 245 | 28 | 5.66 | −0.514 | 0/out→out |
| PGSC0003DMG400008649 | PGSC0003DMT400022286* | Ic | StLBD11-3 | 2 | chr11 | 171 | 19 | 6.94 | −0.371 | 0/out→out |
| PGSC0003DMG400027372 | PGSC0003DMT400070405* | Ic | StLBD11-1 | 1 | chr11 | 94 | 11 | 4.16 | −0.264 | 0/out→out |
| PGSC0003DMG400025384 | PGSC0003DMT400065297* | Ic | StLBD11-2 | 2 | chr11 | 216 | 23 | 5.05 | −0.219 | 0/out→out |
| PGSC0003DMG402023851 | PGSC0003DMT400061283* | Ic | StLBD12-1 | 2 | chr12 | 169 | 19 | 9.01 | −0.295 | 0/out→out |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Ubi3 | TCCGACACCATCGACAATGT | CGACCATCCTCAAGCTGCTT |
| StLBD1-5 | CAATGCCACTGTCTTCGTCG | CTCCTTTAAGCACTGTTTCTACCG |
| StLBD2-6 | CGAACAGTAAACCCAGTGAACG | TGATCGAGATCCGCCAACAT |
| StLBD3-1 | CGGTTCCTTCGTCGTAAATGTC | GCACGCATGTCTGCCTCATA |
| StLBD3-2 | CAACGAGGCTGAGGTGAGACTTA | TGACCAGAAGTGTCCGCAAAA |
| StLBD3-5 | CTTGGACCTGACCATTTGCG | GGGAGATCGGGATCAGAGTTATTT |
| StLBD6-5 | CCTGAATGTTATTAGAGCGGAGAT | CGAGGTGGTTGTAGACGGTTG |
| StLBD11-2 | CGCTAAGGCACAAGCTGAAA | ATAGGGTCTCCCATGATCCAAT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Cao, M.; Chen, X.; Ye, M.; Zhao, P.; Nan, Y.; Li, W.; Zhang, C.; Kong, L.; Kong, N.; et al. Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in Solanum tuberosum. Int. J. Mol. Sci. 2019, 20, 5360. https://doi.org/10.3390/ijms20215360
Liu H, Cao M, Chen X, Ye M, Zhao P, Nan Y, Li W, Zhang C, Kong L, Kong N, et al. Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in Solanum tuberosum. International Journal of Molecular Sciences. 2019; 20(21):5360. https://doi.org/10.3390/ijms20215360
Chicago/Turabian StyleLiu, Hengzhi, Minxuan Cao, Xiaoli Chen, Minghui Ye, Peng Zhao, Yunyou Nan, Wan Li, Chao Zhang, Lingshuang Kong, Nana Kong, and et al. 2019. "Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in Solanum tuberosum" International Journal of Molecular Sciences 20, no. 21: 5360. https://doi.org/10.3390/ijms20215360
APA StyleLiu, H., Cao, M., Chen, X., Ye, M., Zhao, P., Nan, Y., Li, W., Zhang, C., Kong, L., Kong, N., Yang, C., Chen, Y., Wang, D., & Chen, Q. (2019). Genome-Wide Analysis of the Lateral Organ Boundaries Domain (LBD) Gene Family in Solanum tuberosum. International Journal of Molecular Sciences, 20(21), 5360. https://doi.org/10.3390/ijms20215360




