Identification of Rice Large Grain Gene GW2 by Whole-Genome Sequencing of a Large Grain-Isogenic Line Integrated with Japonica Native Gene and Its Linkage Relationship with the Co-integrated Semidwarf Gene d60 on Chromosome 2
Abstract
:1. Introduction
2. Results
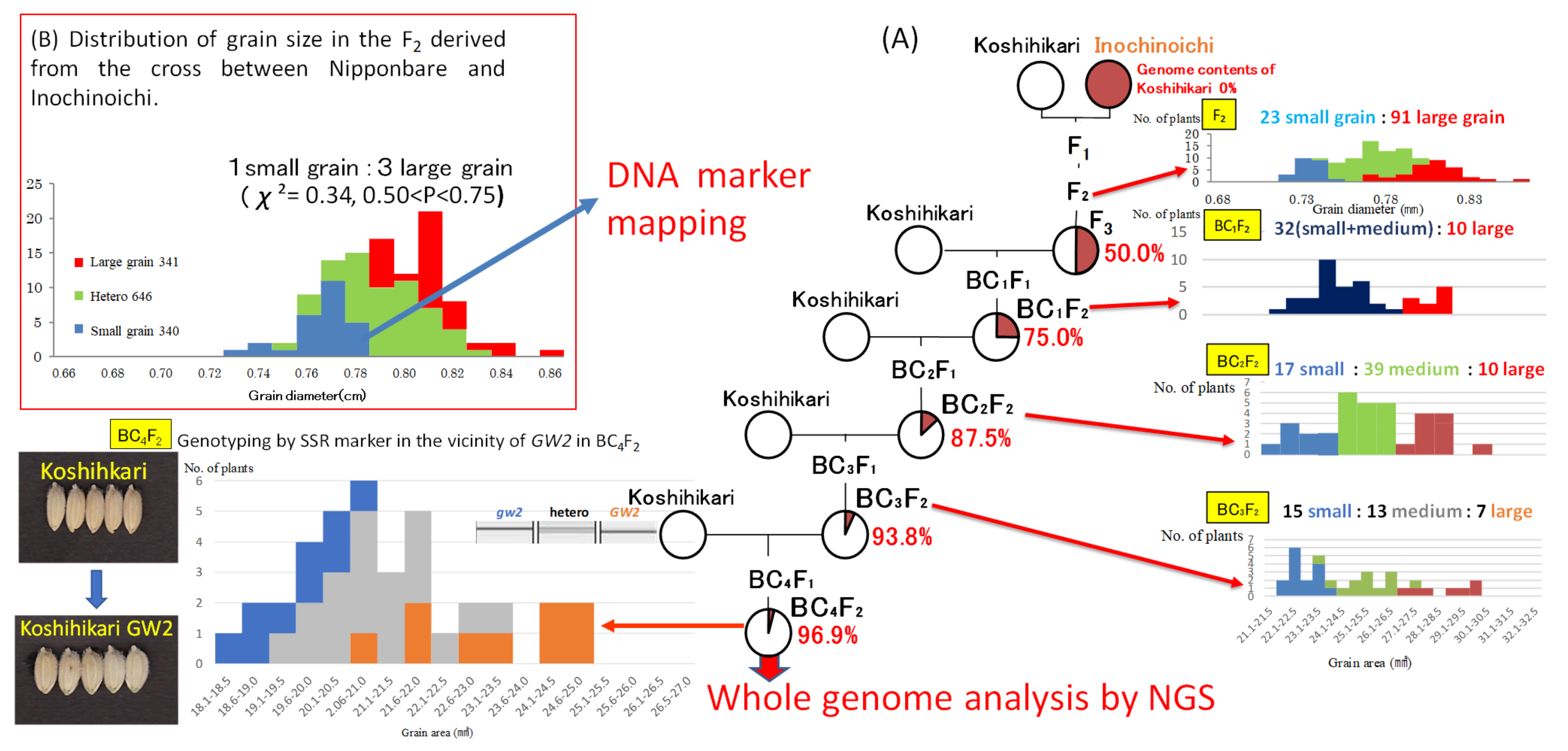
2.1. Inheritance and Phenotypic Expression of Large Grain Gene in an Isogenic Background
2.2. Candidate Region of Large Grain Gene
2.3. Identification of DNA Variation Responsible for Large Grain Using Next-Generation Sequencing (NGS)
2.4. Linkage Relationship Between Semidwarfing Gene d60 and Large Grain Gene GW2
3. Discussion
4. Materials and Methods
4.1. Genetic Analysis
4.2. Mapping of Large Grain Gene by DNA Markers
4.3. Next-generation Sequencing (NGS) Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khush, G.S. Green revolution: Preparing for the 21st century. Genome 1999, 42, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, B.S.; Jabran, K.; Mahajan, G. Rice Production Worldwide; Springer International Publishing AG: Cham, Switzerland, 2017; p. 547. [Google Scholar]
- Fan, C.; Xing, Y.; Man, H.; Lu, T.; Han, B.; Xu, C.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nature Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nature Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nature Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nature Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nature Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Japan Ministry of Agriculture, Forestry and Fisheries. TPP related information. Available online: http://www.maff.go.jp/j/kanbo/tpp/ (accessed on 19 September 2019).
- Japan Meteorological Agency. Heisei 30 July 7 heavy rain and floods. Available online: https://www.data.jma.go.jp/obd/stats/data/bosai/report/2018/20180713/20180713.html (accessed on 19 September 2019).
- Japan Meteorological Agency. 2018 (Heisei 30) Typhoons (Quick Report). Available online: https://www.jma.go.jp/jma/press/1812/21f/typhoon2018.pdf (accessed on 19 September 2019).
- Japan Ministry of Agriculture, Forestry and Fisheries. Damage situation by Heisei 30 July 7 heavy rain and floods. Available online: http://www.maff.go.jp/j/saigai/ooame/20180628.html (accessed on 19 September 2019).
- Japan Meteorological Agency. Abnormal Weather Risk Map. Available online: http://www.data.jma.go.jp/cpdinfo/riskmap/heavyrain.html (accessed on 19 September 2019).
- Japan Meteorological Agency. Long-term changes in the number and frequency of short-term heavy rains observed at AMeDAS. Available online: http://www.jma.go.jp/jma/kishou/info/heavyraintrend.html (accessed on 19 September 2019).
- Tomita, M. Combining two semidwarfing genes d60 and sd1 for reduced height in ‘Minihikari’, a new rice germplasm in the ‘Koshihikari’ genetic background. Genet. Res. Camb. 2012, 94, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant. Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.S.; Kim, Y.J.; Markkandan, K.; Koo, Y.J.; Song, J.T.; Seo, H.S. GW2 functions as an E3 Ubiquitin ligase for rice Expansin-Like 1. Int. J. Mol. Sci. 2018, 19, 1904. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Saito, K.; Yamazaki, K.; Mikami, T. Environmental response of yielding capacity in isogenic lines for grain size of rice. Japan. J. Breed. 1987, 37, 309–317. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweency, M.; Matsumoto, T.; Kanamori, H.; Padhukasaharam, B.; Bustamante, C.; Yoshimura, A.; Doi, K.; et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Official Gazette No. 7080, 14 Aug. 2017, Ministry of Agriculture, Forestry and Fisheries. Plant varietal registration application No. 32364, “Koshihaikri Suruga Gg” 2017, Tokyo, Japan. Available online: http://www.hinshu2.maff.go.jp/vips/cmm/apCMM111.aspx?SHUTSUGAN_NO=32364&LANGUAGE=English (accessed on 14 August 2017).
- Official Gazette No. 7080, 14 Aug. 2017, Ministry of Agriculture, Forestry and Fisheries. Plant varietal registration application No. 32365, “Koshihikari Suruga d60Gg” 2017, Tokyo, Japan. Available online: http://www.hinshu2.maff.go.jp/vips/cmm/apCMM111.aspx?SHUTSUGAN_NO=32365&LANGUAGE=English (accessed on 14 August 2017).
- Mishra, R.; Joshi, R.K.; Zhao, K. Genome editing in rice: Recent advances, challenges, and future implications. Front Plant Sci. 2018, 9, 1361. [Google Scholar] [CrossRef] [PubMed]




| GW2 Koshihikari | GW2 + d60 Koshihikari | Nigata Koshihikari | ||
|---|---|---|---|---|
| Stem length (cm) | 92 | 75 | 99 | |
| Weight of unpolished rice/1000 grains (g) | 29.6 (×1.34) | 28.8 (×1.31) | 22.0 | |
| Polished rice | Taste value | 80.0 | 80.0 | 81.0 |
| Protein | 6.4 | 6.1 | 6.0 | |
| Moisture | 14.4 | 14.5 | 14.5 | |
| Amylose | 18.8 | 18.6 | 18.5 | |
| Normal grain size | 94.7 | 95.6 | 97.3 | |
| Powdery grain | 2.9 | 2.1 | 1.8 | |
| Damaged grain | 0.0 | 0.0 | 0.0 | |
| Colored grain | 0.0 | 0.0 | 0.0 | |
| Split grain | 0.2 | 0.3 | 0.3 | |
| Crashed grain | 2.2 | 2.0 | 0.6 | |
| White degree | 42.6 | 42.8 | 44.8 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomita, M.; Yazawa, S.; Uenishi, Y. Identification of Rice Large Grain Gene GW2 by Whole-Genome Sequencing of a Large Grain-Isogenic Line Integrated with Japonica Native Gene and Its Linkage Relationship with the Co-integrated Semidwarf Gene d60 on Chromosome 2. Int. J. Mol. Sci. 2019, 20, 5442. https://doi.org/10.3390/ijms20215442
Tomita M, Yazawa S, Uenishi Y. Identification of Rice Large Grain Gene GW2 by Whole-Genome Sequencing of a Large Grain-Isogenic Line Integrated with Japonica Native Gene and Its Linkage Relationship with the Co-integrated Semidwarf Gene d60 on Chromosome 2. International Journal of Molecular Sciences. 2019; 20(21):5442. https://doi.org/10.3390/ijms20215442
Chicago/Turabian StyleTomita, Motonori, Shiho Yazawa, and Yoshimasa Uenishi. 2019. "Identification of Rice Large Grain Gene GW2 by Whole-Genome Sequencing of a Large Grain-Isogenic Line Integrated with Japonica Native Gene and Its Linkage Relationship with the Co-integrated Semidwarf Gene d60 on Chromosome 2" International Journal of Molecular Sciences 20, no. 21: 5442. https://doi.org/10.3390/ijms20215442
APA StyleTomita, M., Yazawa, S., & Uenishi, Y. (2019). Identification of Rice Large Grain Gene GW2 by Whole-Genome Sequencing of a Large Grain-Isogenic Line Integrated with Japonica Native Gene and Its Linkage Relationship with the Co-integrated Semidwarf Gene d60 on Chromosome 2. International Journal of Molecular Sciences, 20(21), 5442. https://doi.org/10.3390/ijms20215442






