A Meta-Analysis of Comparative Transcriptomic Data Reveals a Set of Key Genes Involved in the Tolerance to Abiotic Stresses in Rice
Abstract
:1. Introduction
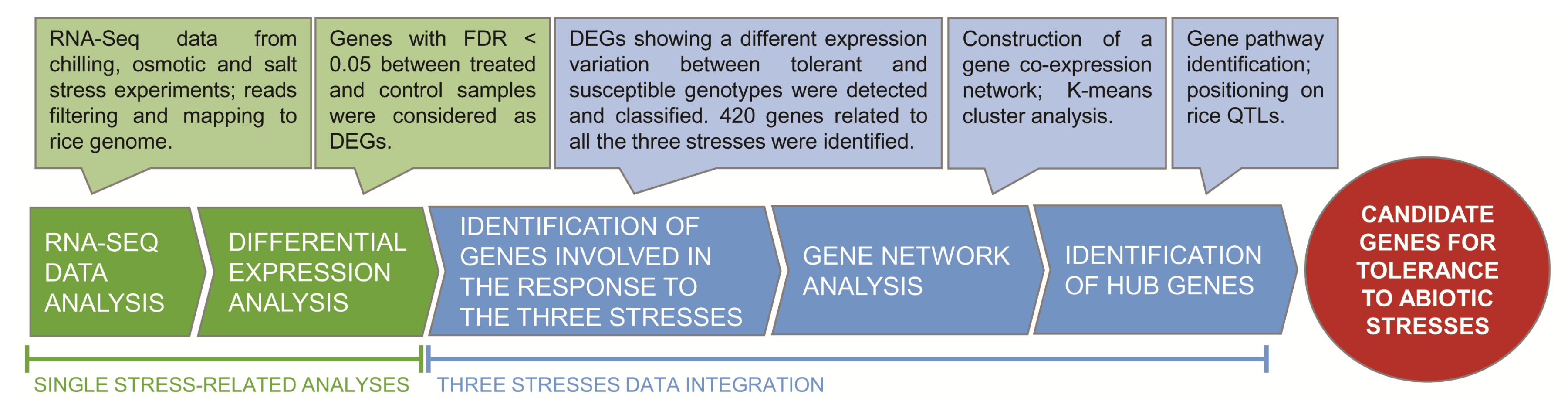
2. Results and Discussion
2.1. RNA-Seq Data Processing
2.2. Differential Gene Expression in Response to Each Single Stress
2.3. Identification of Genes Differentiating the Response of Tolerant and Susceptible Genotypes
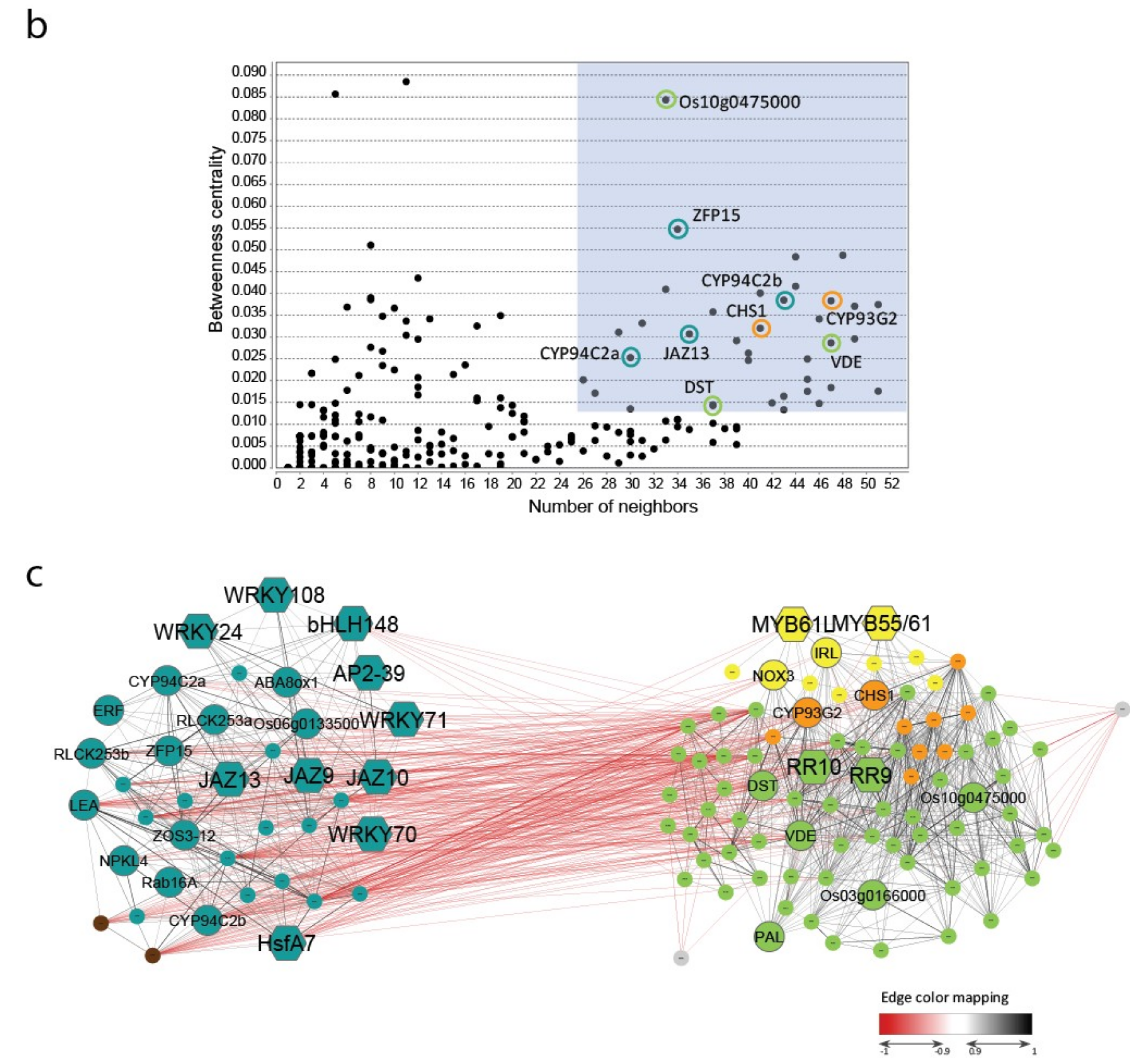
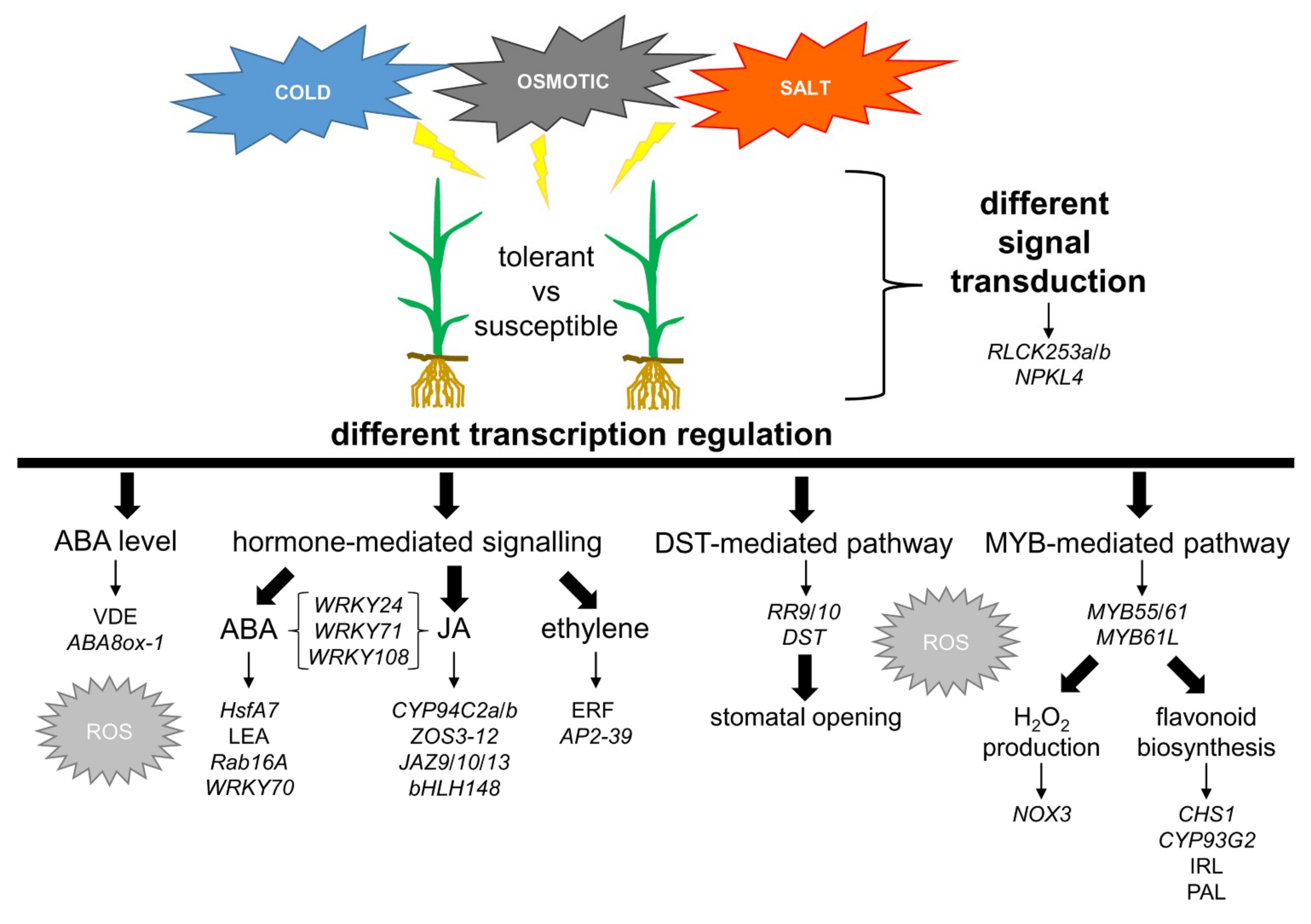
2.4. Characterization of Genes and Pathways Putatively Involved in Abiotic Stress Tolerance
2.5. ABA Synthesis and Metabolism
2.6. The ABA-Mediated Response Pathway and the Crosstalk with Other Signalling Pathways
2.7. The JA-Mediated Response Pathway
2.8. The DST-Related Pathway
2.9. MYBs-Guided Subnetwork Interacts with Flavonoid Biosynthesis and ROS Response
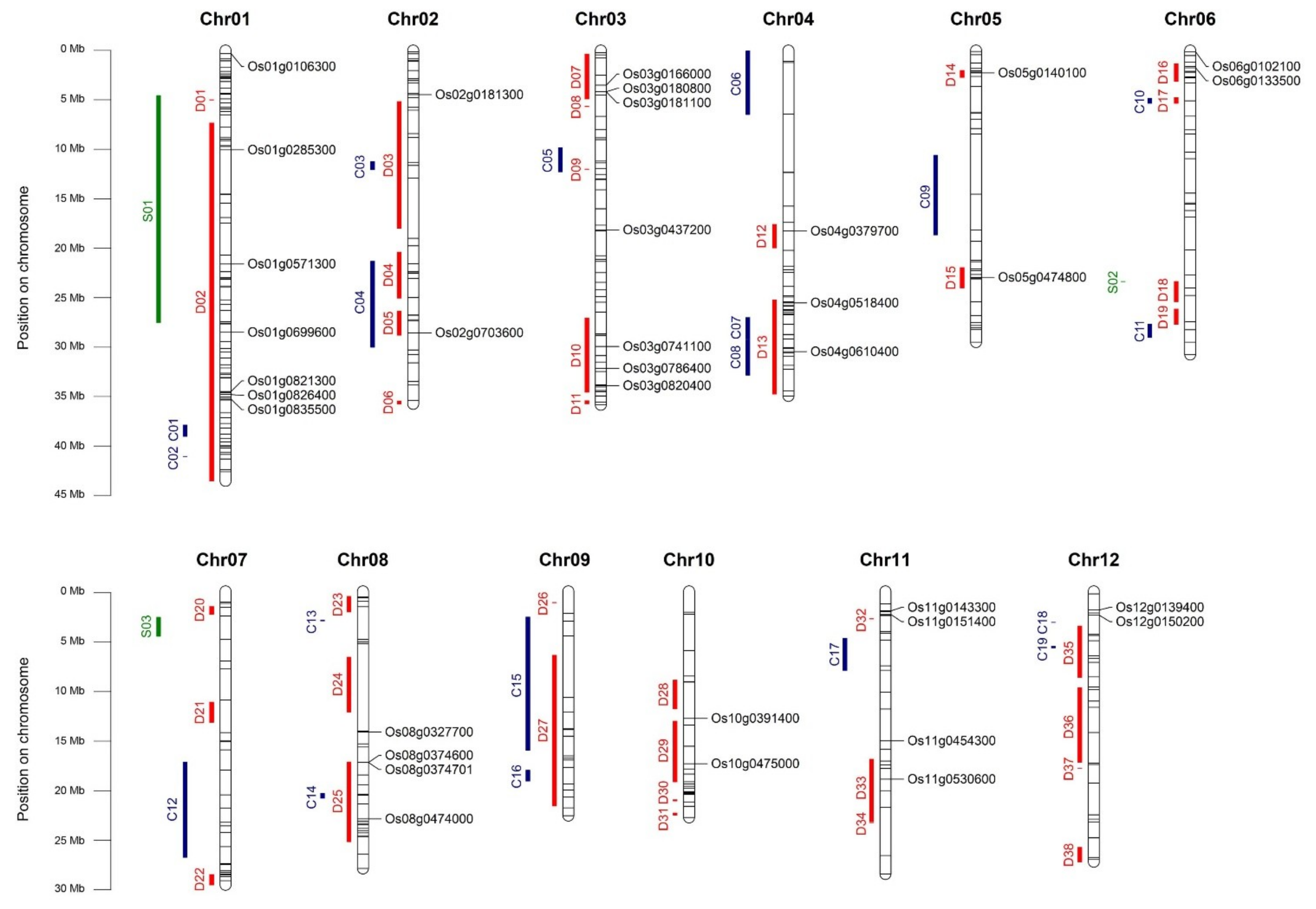
2.10. HUB Genes Distribution on Genome and Co-Localization with Stress-Related QTL
3. Materials and Methods
3.1. Transcriptome Data
3.2. RNA-Seq Data Handling and Mapping to Rice Genome
3.3. Differential Expression and GO-Enrichment Analyses
3.4. Correlation, Network and Clustering Analyses
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, Y.; Jahan, N.; Chen, G.; Ren, D.; Guo, L. Sensing of abiotic stress and ionic stress responses in plants. Int. J. Mol. Sci. 2018, 19, 3298. [Google Scholar] [CrossRef] [PubMed]
- Giordani, T.; Buti, M.; Natali, L.; Pugliesi, C.; Cattonaro, F.; Morgante, M.; Cavallini, A. An analysis of sequence variability in eight genes putatively involved in drought response in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2011, 122, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- da Maia, L.C.; Cadore, P.R.B.; Benitez, L.C.; Danielowski, R.; Braga, E.J.B.; Fagundes, P.R.R.; Magalhães, A.M.; Costa de Oliveira, A. Transcriptome profiling of rice seedlings under cold stress. Funct. Plant Biol. 2016, 44, 419–430. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Tansley insight Salinity tolerance of crops—What is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Buti, M.; Giordani, T.; Vukich, M.; Gentzbittel, L.; Pistelli, L.; Cattonaro, F.; Morgante, M.; Cavallini, A.; Natali, L. HACRE1, a recently inserted copia-like retrotransposon of sunflower (Helianthus annuus L.). Genome 2009, 52, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, J.; Chen, L.; Huang, X.; Cheng, Z.; Han, B.; Zhang, Q.; Wu, C. Rice Functional Genomics Research: Past Decade and Future. Mol. Plant 2018, 11, 359–380. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Kreps, J.A.; Wu, Y.; Chang, H.-S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, M.A.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and cold stress. Plant Physiol. 2003, 133, 1755–1767. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Strnad, M. Jasmonate signaling in plant stress responses and development–active and inactive compounds. New Biotechnol. 2016, 33, 604–613. [Google Scholar] [CrossRef] [PubMed]
- de Ollas, C.; Dodd, I.C. Physiological impacts of ABA–JA interactions under water-limitation. Plant Mol. Biol. 2016, 91, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.; Davies, W.J. Drought, ozone, ABA and ethylene: New insights from cell to plant to community. Plant Cell Environ. 2010, 33, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.-J.; Zhou, Y.-H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.-Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Blumwald, E. The Roles of ROS and ABA in Systemic Acquired Acclimation. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FAO EST: Rice Market Monitor (RMM). Available online: http://www.fao.org/economic/est/publications/rice-publications/rice-market-monitor-rmm/en/ (accessed on 3 September 2018).
- Korres, N.E.; Norsworthy, J.K.; Burgos, N.R.; Oosterhuis, D.M. Temperature and drought impacts on rice production: An agronomic perspective regarding short- and long-term adaptation measures. Water Resour. Rural Dev. 2017, 9, 12–27. [Google Scholar] [CrossRef]
- Radanielson, A.M.; Gaydon, D.S.; Li, T.; Angeles, O.; Roth, C.H. Modeling salinity effect on rice growth and grain yield with ORYZA v3 and APSIM-Oryza. Eur. J. Agron. 2018, 100, 44–45. [Google Scholar] [CrossRef]
- Buti, M.; Pasquariello, M.; Ronga, D.; Milc, J.A.; Pecchioni, N.; Ho, V.T.; Pucciariello, C.; Perata, P.; Francia, E. Transcriptome profiling of short-term response to chilling stress in tolerant and sensitive Oryza sativa ssp. Japonica seedlings. Funct. Integr. Genomics 2018, 6, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Li, D.; He, R.; Fang, Z.; Xia, Y.; Gao, J.; Shen, H.; Cao, M. Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J. Plant Biol. 2014, 57, 337–348. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, X.; Wang, W.; Pan, Y.; Huang, L.; Liu, X.; Zong, Y.; Zhu, L.; Yang, D.; Fu, B. Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS ONE 2012, 7, e43274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baldoni, E.; Bagnaresi, P.; Locatelli, F.; Mattana, M.; Genga, A. Comparative leaf and root transcriptomic analysis of two rice Japonica cultivars reveals major differences in the root early response to osmotic stress. Rice 2016, 9, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degenkolbe, T.; Do, P.T.; Zuther, E.; Repsilber, D.; Walther, D.; Hincha, D.K.; Köhl, K.I. Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Mol. Biol. 2009, 69, 133–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenka, S.K.; Katiyar, A.; Chinnusamy, V.; Bansal, K.C. Comparative analysis of drought-responsive transcriptome in Indica rice genotypes with contrasting drought tolerance. Plant Biotechnol. J. 2011, 9, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Moumeni, A.; Satoh, K.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Kikuchi, S. Transcriptional profiling of the leaves of near-isogenic rice lines with contrasting drought tolerance at the reproductive stage in response to water deficit. BMC Genom. 2015, 16, 1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formentin, E.; Sudiro, C.; Perin, G.; Riccadonna, S.; Barizza, E.; Baldoni, E.; Lavezzo, E.; Stevanato, P.; Sacchi, G.A.; Fontana, P.; et al. Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front. Plant Sci. 2018, 9, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, S.; Zhu, C.; Bai, Z.; Huang, J.; Zhu, L.; Cao, X.; Nanda, S.; Hussain, S.; Riaz, A.; Liang, Q.; et al. iTRAQ-based protein profiling and biochemical analysis of two contrasting rice genotypes revealed their differential responses to salt stress. Int. J. Mol. Sci. 2019, 20, 547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.Y.; Ma, A.; Ramamoorthy, R.; Ramachandran, S. Genome-wide survey on genomic variation, expression divergence, and evolution in two contrasting rice genotypes under high salinity stress. Genome Biol. Evol. 2013, 5, 2032–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, S.; Kaur, N.; Pati, P.K. Reactive oxygen species dynamics in roots of salt sensitive and salt tolerant cultivars of rice. Anal. Biochem. 2018, 550, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Zheng, Y.; Vemireddy, L.R.; Panda, S.K.; Jose, S.; Ranjan, A.; Panda, P.; Govindan, G.; Cui, J.; Wei, K.; et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genom. 2018, 19, 935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankar, R.; Bhattacharjee, A.; Jain, M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 2016, 6, 23719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, M.; Moharana, K.C.; Shankar, R.; Kumari, R.; Garg, R. Genomewide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnol. J. 2014, 12, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.; Dittami, S.M.; Goulitquer, S.; Correa, J.A.; Boyen, C.; Potin, P.; Tonon, T. Transcriptomic and metabolomic analysis of copper stress acclimation in Ectocarpus siliculosus highlights signaling and tolerance mechanisms in brown algae. BMC Plant Biol. 2014, 14, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaar-Moshe, L.; Hübner, S.; Peleg, Z. Identification of conserved drought-adaptive genes using a cross-species meta-analysis approach. BMC Plant Biol. 2015, 15, 111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramu, V.S.; Paramanantham, A.; Ramegowda, V.; Mohan-Raju, B.; Udayakumar, M.; Senthil-Kumar, M.S.K. Transcriptome analysis of sunflower genotypes with contrasting oxidative stress tolerance reveals individual-And combined-biotic and abiotic stress tolerance mechanisms. PLoS ONE 2016, 11, e0157522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rest, J.S.; Wilkins, O.; Yuan, W.; Purugganan, M.D.; Gurevitch, J. Meta-analysis and meta-regression of transcriptomic responses to water stress in Arabidopsis. Plant J. 2016, 85, 548–560. [Google Scholar] [CrossRef] [PubMed]
- Smita, S.; Katiyar, A.; Lenka, S.K.; Dalal, M.; Kumar, A.; Mahtha, S.K.; Yadav, G.; Chinnusamy, V.; Pandey, D.M.; Bansal, K.C. Gene network modules associated with abiotic stress response in tolerant rice genotypes identified by transcriptome meta-analysis. Funct. Integr. Genomics 2019. [Google Scholar] [CrossRef] [PubMed]
- Bjornson, M.; Dandekar, A.; Dehesh, K. Determinants of timing and amplitude in the plant general stress response. J. Integr. Plant Biol. 2016, 58, 119–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formentin, E.; Barizza, E.; Stevanato, P.; Falda, M.; Massa, F.; Tarkowskà, D.; Novák, O.; Lo Schiavo, F. Fast regulation of hormone metabolism contributes to salt tolerance in rice (Oryza sativa spp. Japonica, L.) by inducing specific morpho-physiological responses. Plants 2018, 7, 75. [Google Scholar]
- Formentin, E.; Sudiro, C.; Ronci, M.B.; Locato, V.; Barizza, E.; Stevanato, P.; Ijaz, B.; Zottini, M.; De Gara, L.; Lo Schiavo, F. H2O2 signature and innate antioxidative profile make the difference between sensitivity and tolerance to salt in rice cells. Front. Plant Sci. 2018, 9, 1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serin, E.A.R.; Nijveen, H.; Hilhorst, H.W.M.; Ligterink, W. Learning from co-expression networks: possibilities and challenges. Front. Plant Sci. 2016, 7, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.X.; Thomas, C.E.; Brunak, S. Network biology concepts in complex disease comorbidities. Nat. Rev. Genet. 2016, 17, 615. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.; Jung, I.; Shin, S.-J.; Park, J.; Rhee, S.; Kim, J.-K.; Jung, W.; Kwon, H.-B.; Kim, S. Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice. Front. Plant Sci. 2017, 8, 1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, P.; Cui, F.; Zhang, F.; Luo, X.; Xie, J. Transcriptome analysis of salt stress responsiveness in the seedlings of dongxiang wild rice (Oryza rufipogon Griff.). PLoS ONE 2016, 11, e0146242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, J.; Liu, S.; Hu, H.; Xiong, L. Systematic analysis of NPK1-like genes in rice reveals a stress-inducible gene cluster co-localized with a quantitative trait locus of drought resistance. Mol. Genet. Genomics 2008, 280, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Lu, S.; Wang, M.; Wang, L.; Wang, W.; Cao, F.; Chen, H.; Wang, J.; Zhang, J.; et al. Inhibition of a basal transcription factor 3-like gene Osj10gBTF3 in rice results in significant plant miniaturization and typical pollen abortion. Plant Cell Physiol. 2012, 53, 2073–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Xu, M.; Wang, Y.; Jamil, M. Basal Transcription Factor 3 plays an important role in seed germination and seedling growth of rice. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narsai, R.; Wang, C.; Chen, J.; Wu, J.; Shou, H.; Whelan, J. Antagonistic, overlapping and distinct responses to biotic stress in rice (Oryza sativa) and interactions with abiotic stress. BMC Genom. 2013, 14, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, J.K.; Wardhan, V.; Singh, D.; Chakraborty, S.; Chakraborty, N. Genome-wide identification of the Alba gene family in plants and stress-responsive expression of the rice Alba genes. Genes 2018, 9, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Cao, J.; He, J.; Chen, Q.; Li, X.; Yang, Y. Molecular Mechanism for the regulation of ABA homeostasis during plant development and stress responses. Int. J. Mol. Sci. 2018, 19, 3643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mega, R.; Meguro-Maoka, A.; Endo, A.; Shimosaka, E.; Murayama, S.; Nambara, E.; Seo, M.; Kanno, Y.; Abrams, S.R.; Sato, Y. Sustained low abscisic acid levels increase seedling vigor under cold stress in rice (Oryza sativa L.). Sci. Rep. 2015, 5, 13819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, G.K.; Yamazaki, M.; Kobayashi, M.; Hirochika, R.; Miyao, A.; Hirochika, H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel ostatc gene. Plant Physiol. 2001, 125, 1248–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jahns, P.; Latowski, D.; Strzalka, K. Mechanism and regulation of the violaxanthin cycle: The role of antenna proteins and membrane lipids. Biochim. Biophys. Acta Bioenerg. 2009, 1787, 3–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Gallie, D.R. Violaxanthin de-epoxidase is rate-limiting for non-photochemical quenching under subsaturating light or during chilling in Arabidopsis. Plant Physiol. Biochem. 2012, 58, 66–82. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Gao, S.; Li, B.; Dong, X.C.; Feng, H.L.; Meng, Q.W. Overexpression of violaxanthin de-epoxidase gene alleviates photoinhibition of PSII and PSI in tomato during high light and chilling stress. J. Plant Physiol. 2010, 167, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, S.; Zuo, K.; Luo, L.; Tang, K. Identification of gene modules associated with drought response in rice by network-based analysis. PLoS ONE 2012, 7, e33748. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.L.; Zou, J.; Liu, C.F.; Zhou, X.Y.; Zhang, X.W.; Luo, G.Y.; Chen, X.B. Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep. 2013, 46, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Jiang, Q.; Liu, J.; Zeng, W.; Zeng, Y.; Li, R.; Luo, J. Comparative transcriptome profiling of chilling tolerant rice chromosome segment substitution line in response to early chilling stress. Genes Genom. 2017, 39, 127–141. [Google Scholar] [CrossRef]
- Nylander, M.; Svensson, J.; Palva, E.T.; Welin, B.V. Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- RoyChoudhury, A.; Roy, C.; Sengupta, D.N. Transgenic tobacco plants overexpressing the heterologous lea gene Rab16A from rice during high salt and water deficit display enhanced tolerance to salinity stress. Plant Cell Rep. 2007, 26, 1839–1859. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, M.; Datta, K.; Roychoudhury, A.; Gayen, D.; Sengupta, D.N.; Datta, S.K. Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal. Behav. 2012, 7, 502–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauwels, L.; Goossens, A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell 2011, 23, 3089–3100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in arabidopsis. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, K.-Y.; Park, M.R.; Mohanty, B.; Herath, V.; Xu, F.; Mauleon, R.; Wijaya, E.; Bajic, V.B.; Bruskiewich, R.; de los Reyes, B.G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010, 10, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John Lilly, J.; Subramanian, B. Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativa L.). Plant Sci. 2019, 280, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Shin, M.; Zou, X.; Huang, J.; Ho, T.H.D.; Shen, Q.J. A negative regulator encoded by a rice WRKY gene represses both abscisic acid and gibberellins signaling in aleurone cells. Plant Mol. Biol. 2009, 70, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gu, L.; Ringler, P.; Smith, S.; Rushton, P.J.; Shen, Q.J. Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 2015, 236, 214–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, L.; Zhang, J.; Zhang, H.; Zhang, Z.; Quan, R.; Zhou, S.; Huang, R. Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS ONE 2011, 6, e25216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Moumeni, A.; Attia, K.; Venuprasad, R.; Serraj, R.; Kumar, A.; Leung, H.; Islam, A.K.M.R.; et al. Comparative transcriptome analysis of AP2/EREBP gene family under normal and hormone treatments, and under two drought stresses in NILs setup by Aday Selection and IR64. Mol. Genet. Genomics 2012, 287, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.Y.; Vo, K.T.X.; Nguyen, C.D.; Jeong, D.H.; Lee, S.K.; Kumar, M.; Kim, S.R.; Park, S.H.; Kim, J.K.; Jeon, J.S. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71. Plant Biotechnol. Rep. 2016, 10, 13–23. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, S.; Yang, T.; Zeng, Z.; Huang, Z.; Liu, Q.; Wang, X.; Leach, J.; Leung, H.; Liu, B. Global transcriptional profiling of a cold-tolerant rice variety under moderate cold stress reveals different cold stress response mechanisms. Physiol. Plant. 2015, 154, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Cheng, D.; Yang, Y.; Zhang, G.; Qin, M.; Chen, J.; Chen, Y.; Jiang, M. Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol. 2017, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Kurotani, K.I.; Hayashi, K.; Hatanaka, S.; Toda, Y.; Ogawa, D.; Ichikawa, H.; Ishimaru, Y.; Tashita, R.; Suzuki, T.; Ueda, M.; et al. Elevated levels of CYP94 family gene expression alleviate the Jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 2015, 56, 779–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- To, H.T.M.; Nguyen, H.T.; Dang, N.T.M.; Nguyen, N.H.; Bui, T.X.; Lavarenne, J.; Phung, N.T.P.; Gantet, P.; Lebrun, M.; Bellafiore, S.; et al. Unraveling the genetic elements involved in shoot and root growth regulation by jasmonate in rice using a genome-wide association study. Rice 2019, 12, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peethambaran, P.K.; Glenz, R.; Höninger, S.; Shahinul Islam, S.M.; Hummel, S.; Harter, K.; Kolukisaoglu, Ü.; Meynard, D.; Guiderdoni, E.; Nick, P.; et al. Salt-inducible expression of OsJAZ8 improves resilience against salt-stress. BMC Plant Biol. 2018, 18, 311. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Du, H.; Tang, N.; Li, X.; Xiong, L. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Kawahara-Miki, R.; Miyamoto, K.; Yamane, H.; Nojiri, H.; Tsujii, Y.; Okada, K. OsMYC2 mediates numerous defence-related transcriptional changes via jasmonic acid signalling in rice. Biochem. Biophys. Res. Commun. 2017, 486, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ye, H.; Yao, R.; Zhang, T.; Xiong, L. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.; Ma, S.; Li, X.; Xiong, L. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y. Do OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-Y.; Chao, D.-Y.; Gao, J.-P.; Zhu, M.-Z.; Shi, M.; Lin, H.-X. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 2009, 23, 1805–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet. 2015, 11, e1005617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Zhao, B.; Yuan, D.; Duan, M.; Qian, Q.; Tang, L.; Wang, B.; Liu, X.; Zhang, J.; Wang, J.; et al. Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. USA 2013, 110, 3167–3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.; Jiao, F.; Chu, J.; Jin, G.; Chen, M.; Wu, P. The two-component signal system in rice (Oryza sativa L.): A genome-wide study of cytokinin signal perception and transduction. Genomics 2007, 89, 697–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imamura, A.; Hanaki, N.; Nakamura, A.; Suzuki, T.; Taniguchi, M.; Kiba, T.; Ueguchi, C.; Sugiyama, T.; Mizuno, T. Compilation and characterization of Arabiopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999, 40, 733–742. [Google Scholar] [CrossRef] [PubMed]
- Pavlů, J.; Novák, J.; Koukalová, V.; Luklová, M.; Brzobohatý, B.; Černý, M. Cytokinin at the crossroads of abiotic stress signalling pathways. Int. J. Mol. Sci. 2018, 19, 2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, D.W.; Mok, M.C. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors; active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB Repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Wu, K.; Chen, J.; Liu, Q.; Wu, Y.; Liu, B.; Fu, X. OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 2018, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nutan, K.K.; Kushwaha, H.R.; Singla-Pareek, S.L.; Pareek, A. Transcription dynamics of Saltol QTL localized genes encoding transcription factors, reveals their differential regulation in contrasting genotypes of rice. Funct. Integr. Genom. 2017, 17, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K.; Kondo, M.; Aya, K.; Miyao, A.; Sato, Y.; Antonio, B.A.; Namiki, N.; Nagamura, Y.; Matsuoka, M. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013, 54, 1791–1802. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Park, H.L.; Lee, S.W.; Jung, K.H.; Hahn, T.R.; Cho, M.H. Transcriptomic analysis of UV-treated rice leaves reveals UV-induced phytoalexin biosynthetic pathways and their regulatory networks in rice. Phytochemistry 2013, 96, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Walia, H.; Wilson, C.; Condamine, P.; Liu, X.; Ismail, A.M.; Zeng, L.; Wanamaker, S.I.; Mandal, J.; Xu, J.; Cui, X.; et al. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol. 2005, 139, 822–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Chu, H.; Chu, I.K.; Lo, C. CYP93G2 is a flavanone 2-hydroxylase required for C -glycosylflavone biosynthesis in rice. Plant Physiol. 2010, 154, 324–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markham, K.R.; Tanner, G.J.; Caasi-Lit, M.; Whitecross, M.I.; Nayudu, M.; Mitchell, K.A. Possible protective role for 3’,4’-dihydroxyflavones induced by enhanced UV-B in a UV-tolerant rice cultivar. Phytochemistry 1998, 49, 1913–1919. [Google Scholar] [CrossRef]
- Dong, X.; Chen, W.; Wang, W.; Zhang, H.; Liu, X.; Luo, J. Comprehensive profiling and natural variation of flavonoids in rice. J. Integr. Plant Biol. 2014, 56, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Kim, S.T.; Wang, Y.; Kim, S.K.; Lee, C.H.; Kim, K.K.; Kim, J.K.; Lee, S.Y.; Kang, K.Y. Overexpression of rice isoflavone reductase-like gene (OsIRL) confers tolerance to reactive oxygen species. Physiol. Plant. 2010, 138, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.-M.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.F.; Li, W.Q.; Li, W.Y.; Wu, G.L.; Zhou, C.Y.; Chen, K.M. Characterization of rice NADPH oxidase genes and their expression under various environmental conditions. Int. J. Mol. Sci. 2013, 14, 9440–9458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijaz, B.; Formentin, E.; Ronci, B.; Locato, V.; Barizza, E.; Hyder, M.Z.; Lo Schiavo, F.; Yasmin, T. Salt tolerance in indica rice cell cultures depends on a fine tuning of ROS signalling and homeostasis. PLoS ONE 2019, 14, e0213986. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, F.; Zhao, Q.; Liu, J.; Cheng, F. Involvement of NADPH oxidase isoforms in the production of O2? manipulated by ABA in the senescing leaves of early-senescence-leaf (esl) mutant rice (Oryza sativa). PLoS ONE 2018, 13, e0190161. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ding, Z.; Tie, W.; Yan, Y.; Liu, Y.; Wu, C.; Liu, J.; Wang, J.; Peng, M.; Xu, B.; et al. Comparative physiological and transcriptomic analyses provide integrated insight into osmotic, cold, and salt stress tolerance mechanisms in banana. Sci. Rep. 2017, 7, 43007. [Google Scholar] [CrossRef] [PubMed]
- Yonemaru, J.; Yamamoto, T.; Fukuoka, S.; Uga, Y.; Hori, K.; Yano, M. Q-TARO: QTL Annotation Rice Online Database. Rice 2010, 3, 194–203. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. Babraham Bioinformatics-FastQC a quality control tool for high throughput sequence data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 30 January 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, T.; Simko, V. Visualization of a correlation matrix: Package ‘corrplot’. Statistician 2017, 56, 316–324. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Hartigan, J.A. Clustering. Annu. Rev. Biophys. Bioeng. 1973, 2, 81–102. [Google Scholar] [CrossRef] [PubMed]
- Calinski, T.; Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. Theory Methods 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Lisboa, P.J.G.; Etchells, T.A.; Jarman, I.H.; Chambers, S.J. Finding reproducible cluster partitions for the k-means algorithm. BMC Bioinform. 2013, 14, S8. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Experiment | Sequencing Method | Cultivar | Condition | RNA Sample Names |
|---|---|---|---|---|
| Chilling stress (10 °C, 10 h) | 75 bp paired-end | Thaibonnet chilling susceptible (ChSus) | Control | ChSusC1, ChSusC2, ChSusC3 |
| Treated | ChSusT1, ChSusT2, ChSusT3 | |||
| Volano chilling tolerant (ChTol) | Control | ChTolC1, ChTolC2, ChTolC3 | ||
| Treated | ChTolT1, ChTolT2, ChTolT3 | |||
| Osmotic stress (PEG6000 20%, 24 h) | 50 bp single-end | Loto osmotic stress susceptible (OsSus) | Control | OsSusC1, OsSusC2, OsSusC3 |
| Treated | OsSusT1, OsSusT2, OsSusT3 | |||
| Eurosis osmotic stress tolerant (OsTol) | Control | OsTolC1, OsTolC2, OsTolC3 | ||
| Treated | OsTolT1, OsTolT2, OsTolT3 | |||
| Salt stress (saline solution [NaCl:MgSO4:CaCl2:NaNO2 = 10:2:1:1], 72 h) | 50 bp single-end | Vialone Nano salt susceptible (SaSus) | Control | SaSusC1, SaSusC2, SaSusC3 |
| Treated | SaSusT1, SaSusT2, SaSusT3 | |||
| Baldo salt tolerant (SaTol) | Control | SaTolC1, SaTolC2, SaTolC3 | ||
| Treated | SaTolT1, SaTolT2, SaTolT3 |
| Cultivar Phenotype | Chilling Stress | Osmotic Stress | Salt Stress | |||
|---|---|---|---|---|---|---|
| Susceptible | Tolerant | Susceptible | Tolerant | Susceptible | Tolerant | |
| Cultivar name | ChSus | ChTol | OsSus | OsTol | SaSus | SaTol |
| Active genes | 20,907 | 21,137 | 20,267 | 20,265 | 20,851 | 20,719 |
| Number of DEGs (FDR < 0.05) | 14,944 | 14,355 | 13,722 | 12,654 | 6197 | 1537 |
| Up-regulated (logFC > 0) | 7382 | 7039 | 6692 | 6416 | 3116 | 841 |
| Down-regulated (logFC < 0) | 7562 | 7316 | 7030 | 6238 | 3081 | 696 |
| DEG Class | Chilling | Osmotic | Salt |
|---|---|---|---|
| SusOnly_up | 1321 | 1355 | 2446 |
| SusOnly_down | 1556 | 1927 | 2639 |
| TolOnly_up | 964 | 1083 | 171 |
| TolOnly_down | 1324 | 1131 | 254 |
| ΔLFC > 1 | 425 | 798 | 55 |
| ΔLFC < −1 | 308 | 772 | 75 |
| TOTAL | 5898 | 7066 | 5640 |
| Gene Name | RAP ID | Role in Stress Response | GCN Subgroup | References about Variations in Gene Expression between Contrasting Genotypes | Co-Localization with Known QTLs |
|---|---|---|---|---|---|
| OsZFP15 | Os03g0820400 | unknown function (TF) | A | cold [27] drought [38] | 1 DT (panicle length) |
| OsRLCK253a | Os08g0374600 | signal transduction | A | / | 1 DT (osmotic adjustment) |
| OsRLCK253b | Os08g0374701 | signal transduction | A | / | 1 DT (osmotic adjustment) |
| OsNPKL4 | Os01g0699600 | signal transduction | A | / | 1 DT: qLRC-1 |
| OsABA8ox1 | Os02g0703600 | ABA catabolism | A | osmotic [28] | 1 CT: qSDW2; 2 DT: qGY-2b, qTGW-2a |
| VDE | Os04g0379700 | ABA biosynthesis/xanthophyll cycle | B | / | 1 DT (panicle length) |
| OsHsfA7 | Os01g0571300 | ABA signalling (TF) | A | salt [36] | 1 DT: rfw1b; 1 ST (Na+ uptake) |
| LEA | Os08g0327700 | ABA signalling | A | / | / |
| OsRab16A | Os11g0454300 | ABA signalling | A | salt [36] | / |
| OsWRKY24 | Os01g0826400 | ABA, GA, JA signalling (TF) | A | cold [27] | 1 DT (Panicles/m2) |
| OsWRKY70 | Os05g0474800 | ABA and GA signalling | A | cold [27] salt [36] | 2 DT (panicle or tiller no.per m2, fraction sterile panicles) |
| OsWRKY71 | Os02g0181300 | ABA, GA, JA signalling (TF) | A | cold [27] | / |
| OsWRKY108 | Os01g0821300 | ABA, JA signalling (TF) | A | / | 1 DT (Panicles/m2) |
| OsAP2-39 | Os04g0610400 | ethylene signalling (TF) | A | cold [27] drought [80] | 1 CT: OsAOX1a; 1 DT: rfw4a |
| ERF | Os08g0474000 | ethylene signalling? (TF) | A | cold [27] | 1 DT (osmotic adjustment) |
| OsCYP94C2a | Os11g0151400 | JA inactivation | A | salt [36] | / |
| OsCYP94C2b | Os12g0150200 | JA inactivation | A | / | / |
| OsZOS3-12 | Os03g0437200 | JA signalling (TF) | A | cold [27] | / |
| OsJAZ9 | Os03g0180800 | JA signalling (TF) | A | cold [27] | 1 DT: qtl3.1 |
| OsJAZ10 | Os03g0181100 | JA signalling (TF) | A | cold [27] | 1 DT: qtl3.1 |
| OsJAZ13 | Os10g0391400 | JA signalling (TF) | A | cold [27] salt [36] | / |
| OsbHLH148 | Os03g0741100 | JA signalling (TF) | A | cold [27] | 2 DT (grains per panicle, carbon isotope discrimination) |
| DST | Os03g0786400 | H2O2/CK signalling (TF) | B | / | 1 DT (panicle length) |
| OsRR9 | Os11g0143300 | CK signalling (TF) | B | / | / |
| OsRR10 | Os12g0139400 | CK signalling (TF) | B | / | / |
| OsMYB55-61 | Os01g0285300 | TF | MYB subnet | salt [104] | 1 DT: rfw1b; 1 ST (Na+ uptake) |
| OsMYB61L | Os05g0140100 | TF | MYB subnet | salt [34] | 1 DT (Sterility (%)) |
| OsCHS1 | Os11g0530600 | flavonoid biosynthesis | MYB subnet/B | salt [108] | 3 DT: gpl11.1, gw11.1, yld11.1 |
| OsCYP93G2 | Os06g0102100 | flavonoid biosynthesis | MYB subnet/B | / | / |
| IRL | Os01g0106300 | flavonoid biosynthesis | MYB subnet | salt [33] | / |
| PAL | Os04g0518400 | phenylpropanoid/flavonoid biosynthesis | B | / | 1 DT: rfw4a |
| OsNOX3 | Os01g0835500 | H2O2 signalling | MYB subnet | salt [35] | 1 DT (Panicles/m2) |
| - | Os06g0133500 | unknown function | A | / | 1 DT (leaf rolling score) |
| - | Os03g0166000 | unknown function (TF?) | B | / | 2 DT: rn3, qtl3.1 |
| - | Os10g0475000 | unknown function (alcohol oxidase?) | B | / | 1 DT (Root penetration index) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buti, M.; Baldoni, E.; Formentin, E.; Milc, J.; Frugis, G.; Lo Schiavo, F.; Genga, A.; Francia, E. A Meta-Analysis of Comparative Transcriptomic Data Reveals a Set of Key Genes Involved in the Tolerance to Abiotic Stresses in Rice. Int. J. Mol. Sci. 2019, 20, 5662. https://doi.org/10.3390/ijms20225662
Buti M, Baldoni E, Formentin E, Milc J, Frugis G, Lo Schiavo F, Genga A, Francia E. A Meta-Analysis of Comparative Transcriptomic Data Reveals a Set of Key Genes Involved in the Tolerance to Abiotic Stresses in Rice. International Journal of Molecular Sciences. 2019; 20(22):5662. https://doi.org/10.3390/ijms20225662
Chicago/Turabian StyleButi, Matteo, Elena Baldoni, Elide Formentin, Justyna Milc, Giovanna Frugis, Fiorella Lo Schiavo, Annamaria Genga, and Enrico Francia. 2019. "A Meta-Analysis of Comparative Transcriptomic Data Reveals a Set of Key Genes Involved in the Tolerance to Abiotic Stresses in Rice" International Journal of Molecular Sciences 20, no. 22: 5662. https://doi.org/10.3390/ijms20225662






