Characterizing the Role of TaWRKY13 in Salt Tolerance
Abstract
1. Introduction
2. Results
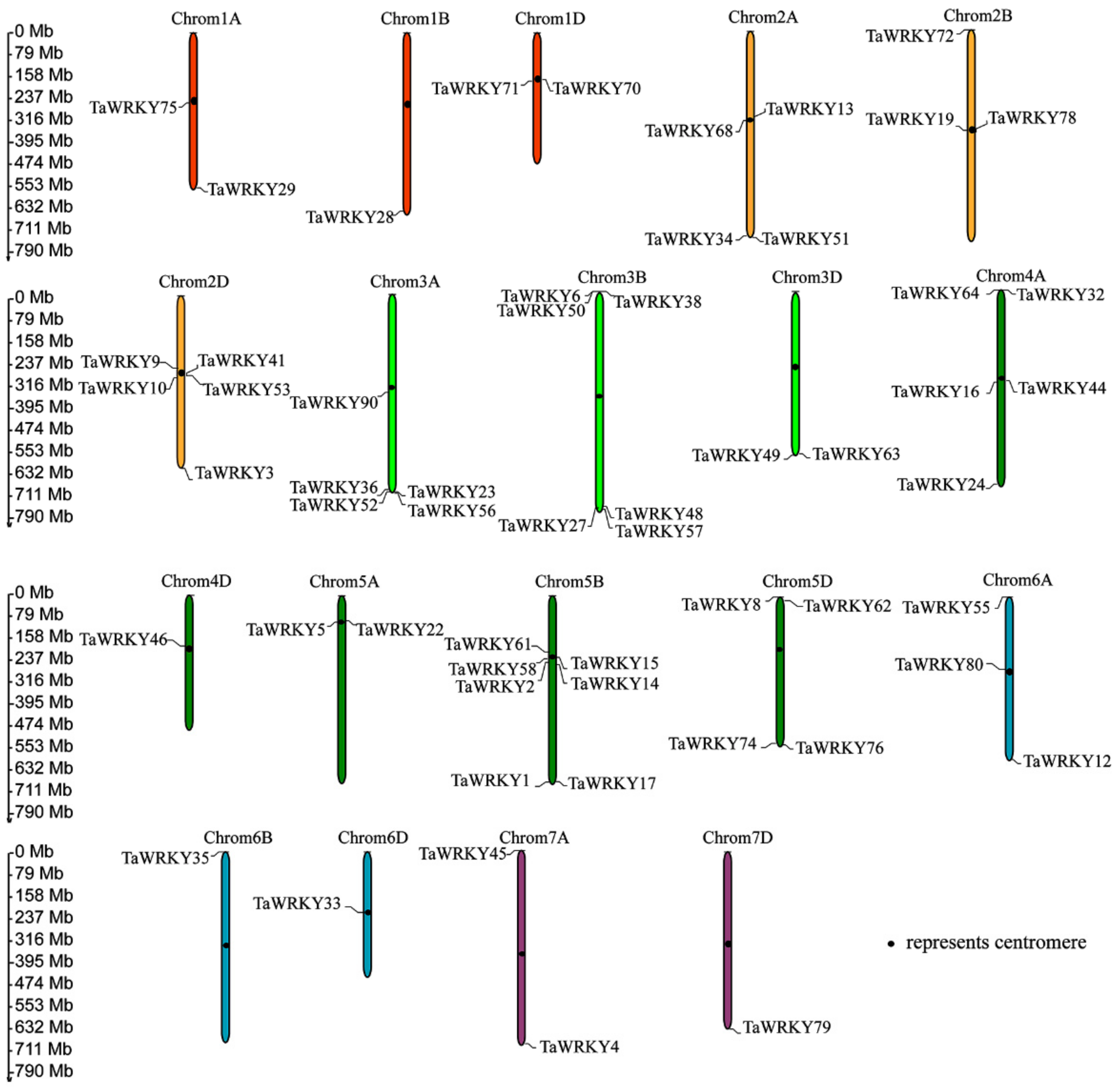
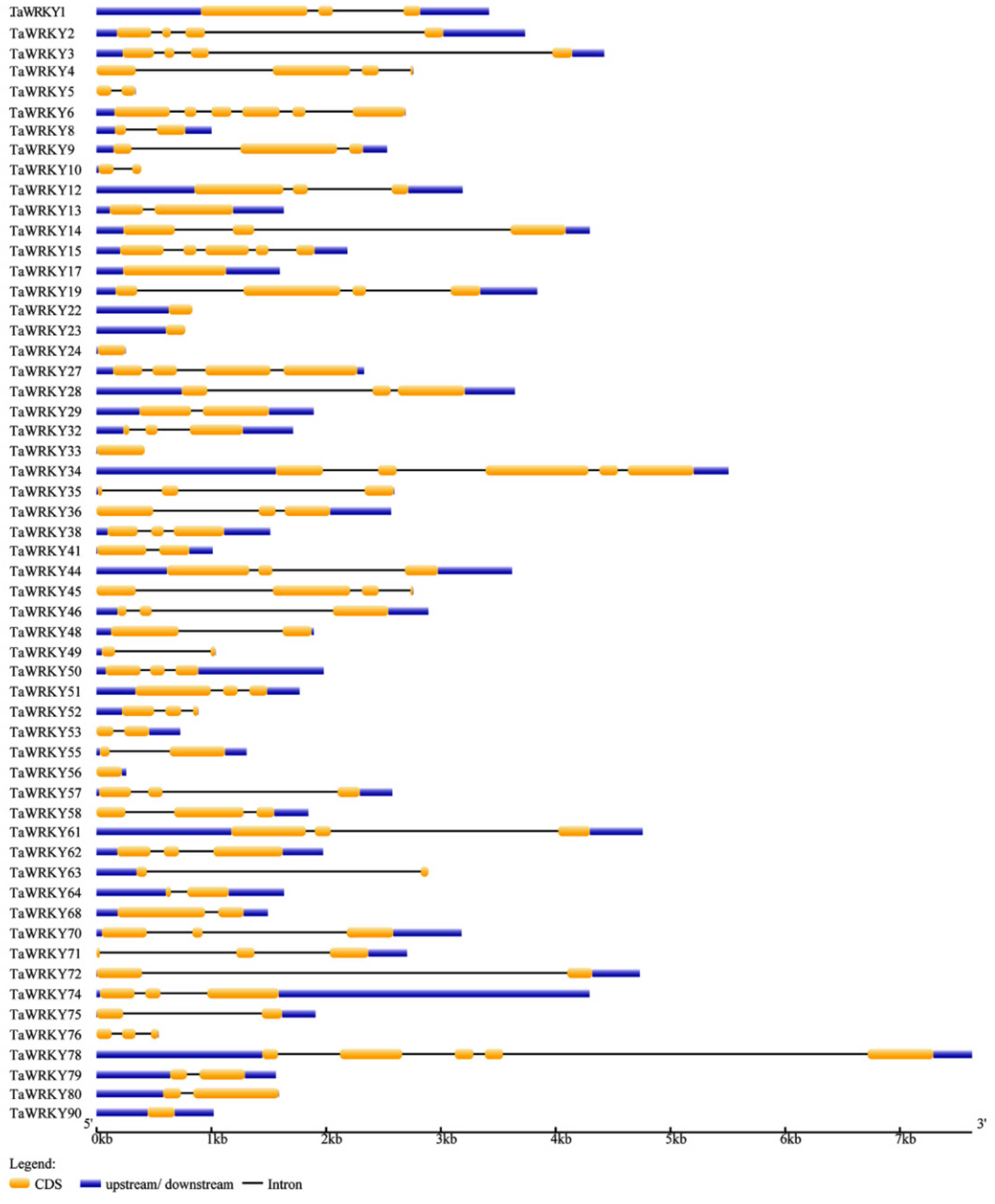
2.1. Identification and Genome Structure Analysis of WRKYs in Triticum aestivum
2.2. Identification and Biological Analysis of TaWRKY13
2.3. Phylogenetic Analysis of AtWRKYs, OsWRKYs and TaWRKYs
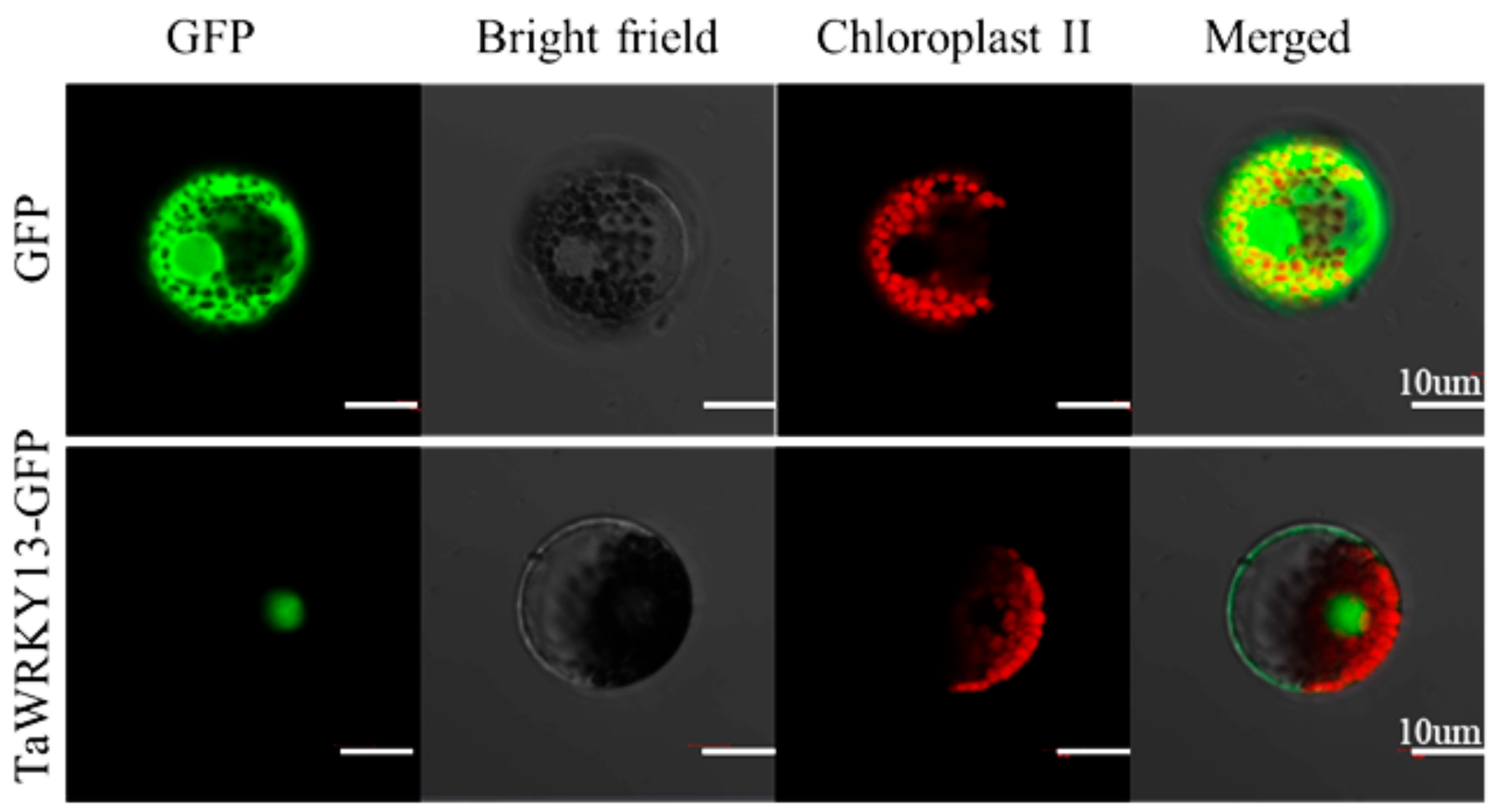
2.4. TaWRKY13 was Localized in the Nucleus
2.5. Tissue-Specific Expression of TaWRKY13
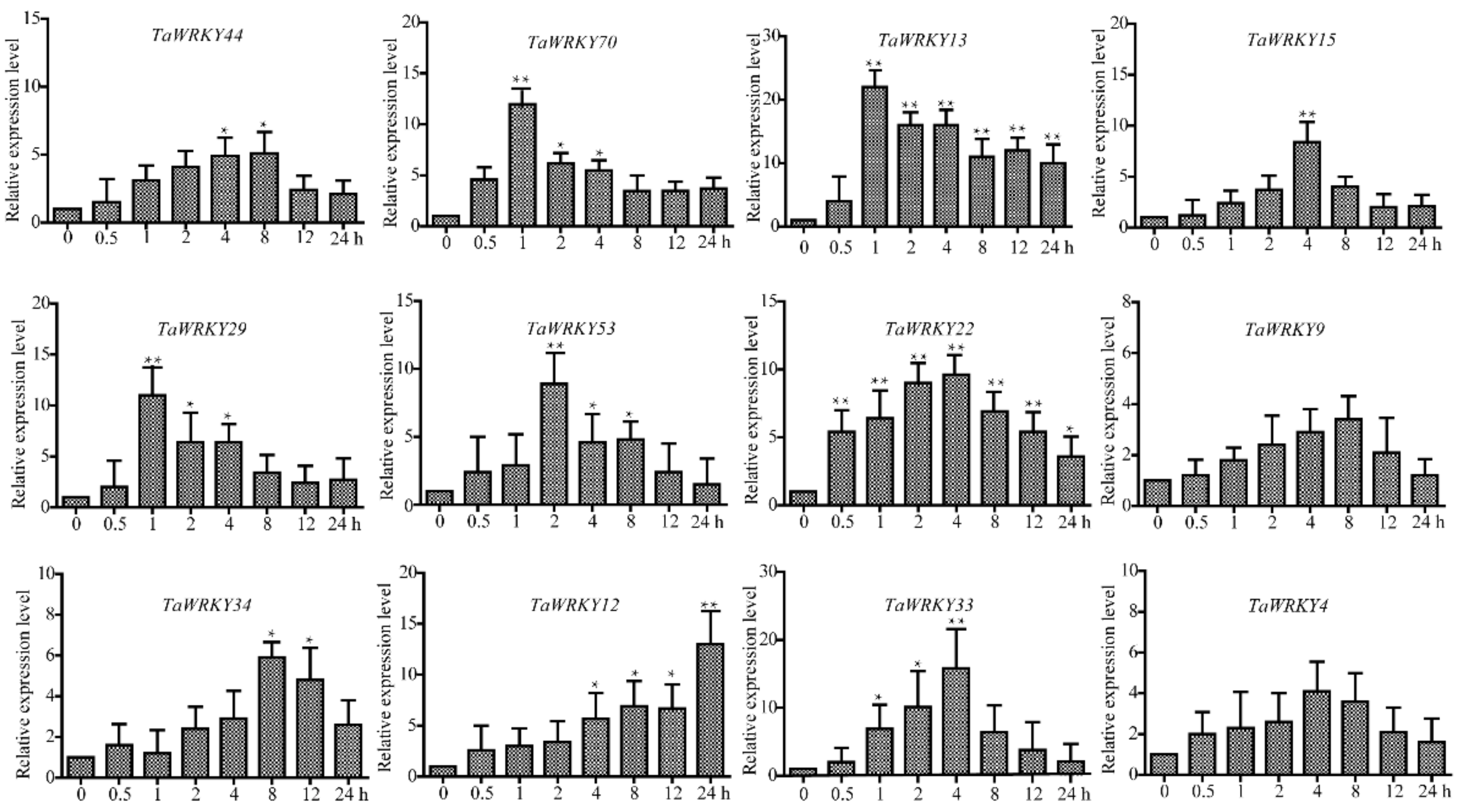
2.6. TaWRKY13 Is Involved in Various Stress Responses
2.7. Stress-Related Regulatory Elements in the Promoter of TaWRKY13
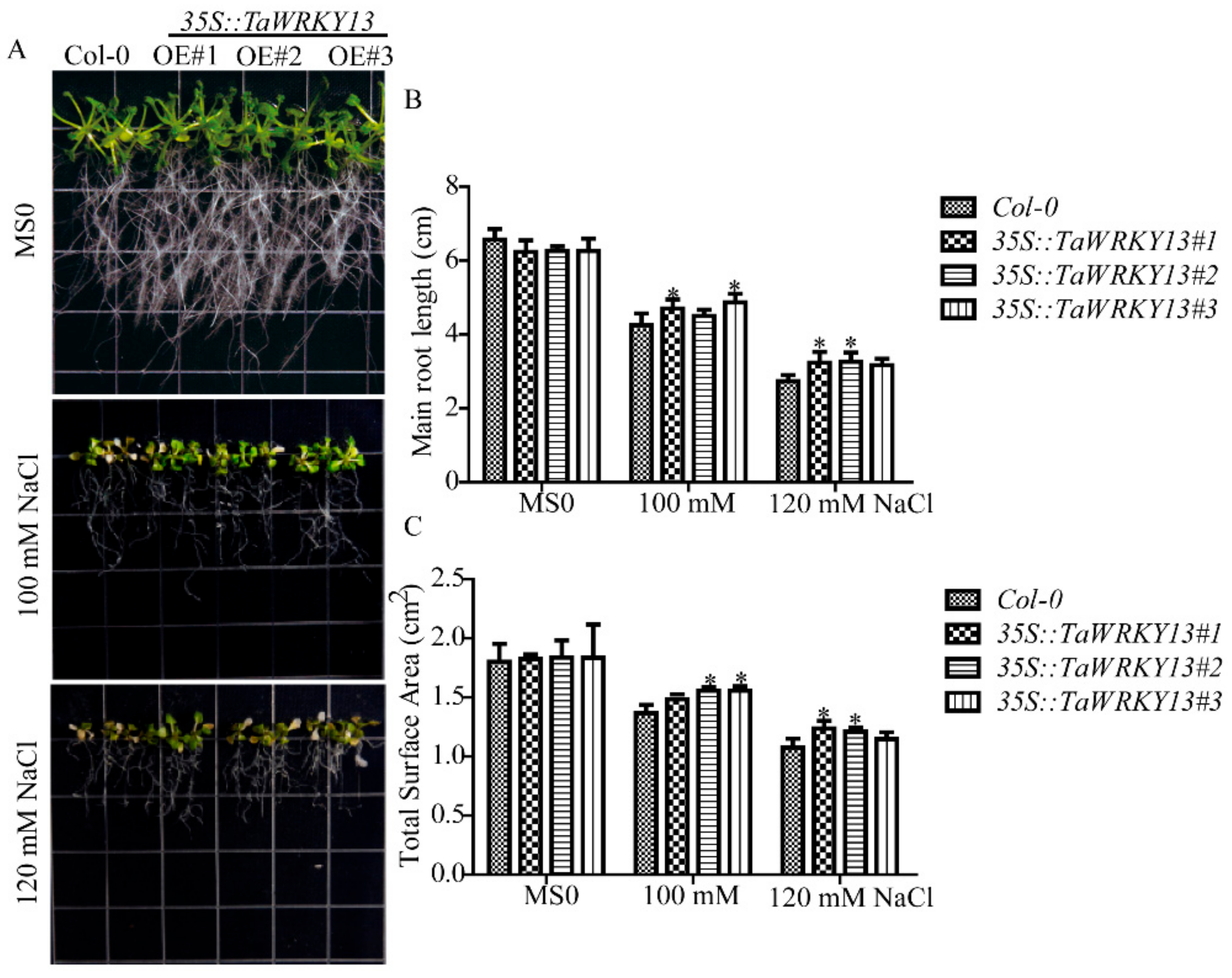
2.8. Root System Analysis Indicates That Overexpression Lines Respond to Salt Stress in Arabidopsis
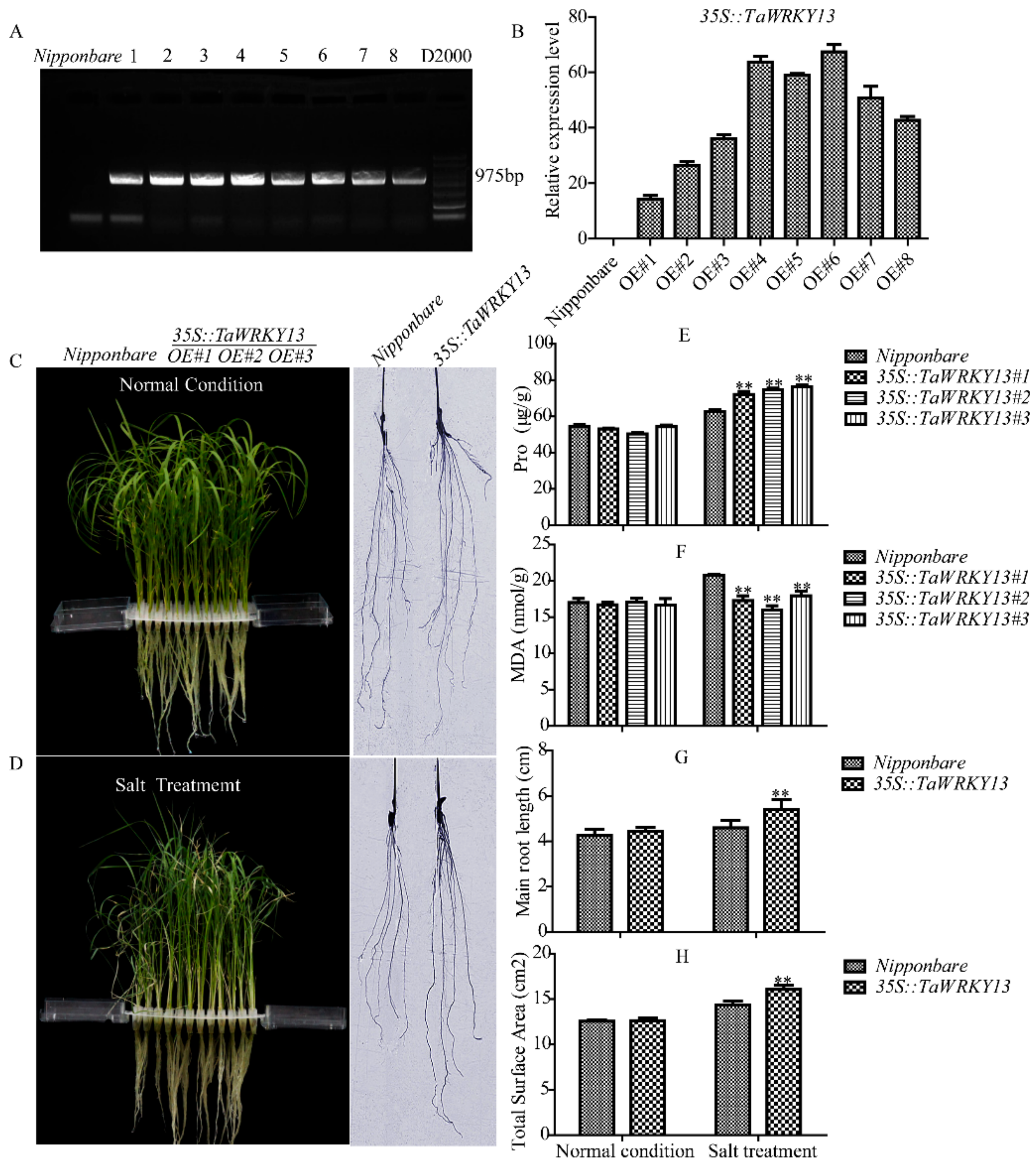
2.9. TaWRKY13 Overexpression Response to Salt Stress in Oryza sativa
3. Discussion
4. Materials and Methods
4.1. De Novo Transcriptome Sequencing of Salt-Treated Wheat
4.2. Identification and Annotation of TaWRKY Response to Salt Stress
4.3. Structure Analysis and Phylogenetic Analysis of TaWRKYs
4.4. RNA Extraction of Stress Treatments and RT-qPCR Analyses
4.5. Gene Isolation and Subcellular Localization
4.6. Tissue-Specific Expression of TaWRKY13 and GUS Staining
4.7. Cis-Acting Elements in the TaWRKY13 Promoter
4.8. Root Growth Assays of TaWRKY13 under Salt Stress in Arabidopsis
4.9. Generation of Transgenic Rice and Stress Identification of TaWRKY13 to Salt Tolerance
4.10. Measurements of Proline (Pro) and Malondialdehyde (MDA) Contents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ROS | Reactive oxygen species |
| SnRK2 | Sucrose non-fermenting 1-related protein kinase 2 |
| MAPK | Mitogen activated protein kinase |
| PKs | Protein kinases |
| TFs | Transcription factors |
| CDS | Coding sequence |
| ORF | Open reading frame |
| SA | Salicylic acid |
| ABA | Abscisic acid |
| ABRE | ABA-responsive element |
| Pro | Proline |
| MDA | Malondialdehyde |
| PEG | Polyethylene glycol |
| GFP | Green fluorescent protein |
| RT-qPCR | Real-time quantitative PCR |
| LTR | Low-temperature responsive |
| GUS | β-glucuronidase |
| X-gluc | 5-bromo-4-chloro-3-indolylb-d-glucuronic acid |
| AGE | Agarose gel electrophoresis |
| MS | Murashige & Skoog |
References
- Bechtold, U.; Field, B. Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 2018, 69, 2753–2758. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 2001, 4, 401–406. [Google Scholar] [CrossRef]
- Bian, S.M.; Jin, D.H.; Li, R.H.; Xie, X.; Gao, G.I.; Sun, W.K.; Li, Y.J.; Zhai, L.L.; Li, X.Y. Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a. Int. J. Mol. Sci. 2017, 18, 2040. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Li, Y.Y.; Liu, B.H.; Kong, F.J.; Tran, L.P. Adaptive Mechanisms of Soybean Grown on Salt-Affected Soils. Land Degrad. Dev. 2017, 29, 1054–1064. [Google Scholar] [CrossRef]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Hao, Y.J. Genome Wide Analysis of the Apple MYB Transcription Factor Family Allows the Identification of MdoMYB121 Gene Conferring Abiotic Stress Tolerance in Plants. PLoS ONE 2013, 8, e69955. [Google Scholar]
- Degregori, J.; Kowalik, T.; Nevins, J.R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 1995, 15, 4215. [Google Scholar] [CrossRef]
- Lei, Y.; Lu, L.; Liu, H.Y.; Li, S.; Xing, F.; Chen, L.L. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 2014, 7, 1494–1496. [Google Scholar] [CrossRef]
- Lang, V.; Palva, E.T. The expression of a Rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. Rep. 1992, 20, 951–962. [Google Scholar] [CrossRef]
- Levine, A.; Tenhaken, R.; Dixon, R.; Lamb, C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 1994, 79, 583–593. [Google Scholar] [CrossRef]
- Michael, S.; Markus, H.; Baldwin, I.T.; Emmanuel, G. Jasmonoyl-L-isoleucine coordinates metabolic networks required for anthesis and floral attractant emission in wild tobacco (Nicotiana attenuata). Plant Cell 2014, 26, 3964–3983. [Google Scholar]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. Cell. Mol. Biol. 2010, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, D. Functions of MAPK Signaling Pathways in the Regulation of Toxicity of Environmental Toxicants or Stresses; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Yoshinaga, K.; Arimura, S.I.; Niwa, Y.; Tsutsumi, N.; Uchimiya, H.; Kawai-Yamada, M. Mitochondrial Behaviour in the Early Stages of ROS Stress Leading to Cell Death in Arabidopsis thaliana. Ann. Bot. 2005, 96, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, W.; Stanley, B.A.; Assmann, S.M. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 2008, 20, 3210–3226. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; He, S.; Gong, W.; Sun, J.; Pan, Z.; Xu, F.; Lu, Y.; Du, X. Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genom. 2014, 15, 760. [Google Scholar] [CrossRef] [PubMed]
- Runnels, L.W.; Yue, L.; Clapham, D.E. TRP-PLIK, a bifunctional protein with kinase and ion channel activities. Science 2001, 291, 1043–1047. [Google Scholar] [CrossRef] [PubMed]
- Mulders, S.M.; Preston, G.M.; Deen, P.M.; Guggino, W.B.; van Os, C.H.; Agre, P. Water channel properties of major intrinsic protein of lens. J. Biol. Chem. 1995, 270, 9010–9016. [Google Scholar] [CrossRef] [PubMed]
- Yovchev, A.G.; Stone, A.K.; Hucl, P.; Scanlon, M.G.; Nickerson, M.T. Polyethylene glycol as an osmotic regulator in dough with reduced salt content. J. Cereal Sci. 2017, 76, 193–198. [Google Scholar] [CrossRef]
- Hunter, T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 1995, 80, 225. [Google Scholar] [CrossRef]
- Madhunita, B.; Ralf, O. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchishinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [PubMed]
- Fiorenza, M.T.; Farkas, T.; Dissing, M.; Kolding, D.; Zimarino, V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995, 23, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchishinozaki, K. NAC transcription factors in plant abiotic stress responses. BBA Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Chen, J.; Nolan, T.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54 and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Response. Plant Cell 2017, 29, 1425. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Cha, J.; Choi, C.; Choi, N.; Ji, H.S.; Park, S.R.; Lee, S.; Hwang, D.J. Rice WRKY11 Plays a Role in Pathogen Defense and Drought Tolerance. Rice 2018, 11, 5. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Bonthala, V.S.; Khandelwal, R.; Jaishankar, J.; Shweta, S.; Nawaz, K.; Prasad, M. Global analysis of WRKY transcription factor superfamily in Setariaidentifies potential candidates involved in abiotic stress signaling. Front. Plant Sci. 2015, 6, 910. [Google Scholar] [CrossRef]
- Shi, W.Y.; Du, Y.T.; Ma, J.; Min, D.H.; Zhang, X.H. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 2018, 19, 4087. [Google Scholar] [CrossRef]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Ding, W.; Fang, W.; Shi, S.; Zhao, Y.; Li, X.; Xiao, K. Wheat WRKY Type Transcription Factor Gene TaWRKY1 is Essential in Mediating Drought Tolerance Associated with an ABA-Dependent Pathway. Plant Mol. Biol. Rep. 2016, 34, 1111–1126. [Google Scholar] [CrossRef]
- Chen, W.; Deng, P.; Chen, L.; Wang, X.; Hui, M.; Wei, H.; Yao, N.; Ying, F.; Chai, R.; Yang, G. A Wheat WRKY Transcription Factor TaWRKY10 Confers Tolerance to Multiple Abiotic Stresses in Transgenic Tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotech. J. 2011, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gao, Y.; Xu, H.; Dai, Y.; Deng, D.; Chen, J. ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 2013, 70, 207–216. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; James, R.A.; Läuchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef] [PubMed]
- Röder, M.S.; Korzun, V.; Wendehake, K.; Plaschke, J.; Tixier, M.; Leroy, P.; Ganal, M.W. A Microsatellite Map of Wheat. Genetics 1998, 149, 2007. [Google Scholar]
- Somers, D.; Isaac, P.; Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theo. Appl. Genet. 2004, 109, 1105–1114. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Martin, T.; Wöhner, R.V.; Hummel, S.; Willmitzer, L.; Frommer, W.B.; Gallagher, S.R. The GUS reporter system as a tool to study plant gene expression. Gus Protocols 1992, 23–43. [Google Scholar] [CrossRef]
- James, D.; Borphukan, B.; Fartyal, D.; Ram, B.; Yadav, R.; Singh, J.; Manna, M.; Panditi, V.; Sheri, V.; Achary, M. Concurrent Overexpression of OsGS1;1 and OsGS2 Genes in Transgenic Rice (Oryza sativa L.): Impact on Tolerance to Abiotic Stresses. Front. Plant Sci. 2018, 9, 786. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Liu, Y.; Shen, Q.J. The WRKY Gene Family in Rice (Oryza sativa). J. Integr. Plant Biol. 2010, 49, 827–842. [Google Scholar] [CrossRef]
- Garnica, D.P.; Upadhyaya, N.M.; Dodds, P.N.; Rathjen, J.P. Strategies for Wheat Stripe Rust Pathogenicity Identified by Transcriptome Sequencing. PLoS ONE 2013, 8, e67150. [Google Scholar] [CrossRef] [PubMed]
- Pingault, L.; Choulet, F.; Alberti, A.; Glover, N.; Wincker, P.; Feuillet, C.; Paux, E. Deep transcriptome sequencing provides new insights into the structural and functional organization of the wheat genome. Genome Biol. 2015, 16, 29. [Google Scholar] [CrossRef]
- Feng, Y.; Zhao, Y.; Wang, K.; Yong, C.L.; Xiang, W.; Yin, J. Identification of vernalization responsive genes in the winter wheat cultivar Jing841 by transcriptome sequencing. J. Genet. 2016, 95, 1–8. [Google Scholar] [CrossRef]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Hao, Y.J.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, Z.B.; Zhang, J.S. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Sezer, O.; Ebru, D.; Turgay, U. Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol. Genet. Genom. 2014, 289, 765–781. [Google Scholar]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Hong, W.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene Expression Profiles during the Initial Phase of Salt Stress in Rice. Plant Cell 2001, 13, 889–905. [Google Scholar] [CrossRef]
- Cavallisforza, L.L.; Edwards, A.W. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967, 21, 550–570. [Google Scholar]
- Chen, J.N.; Yin, Y.H. WRKY Transcription Factors are Involved in Brassinosteroid Signaling and Mediate the Crosstalk Between Plant Growth and Drought Tolerance. Plant Signal. Behav. 2017, 12, e1365212. [Google Scholar] [CrossRef]
- Cheng, H. Overexpression of the GmNAC2 Gene, an NAC Transcription Factor, Reduces Abiotic Stress Tolerance in Tobacco. Plant Mol. Biol. Rep. 2013, 31, 435–442. [Google Scholar]
- Ling, L.D.; Xiao, H.Z.; Chao, Y.P.; Mei, Z.S.; Heng, L.W.; Shu, L.F.; Shu, X.Y. Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genom. 2014, 289, 1103. [Google Scholar]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotech. J. 2010, 6, 486–503. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, N.; Hu, R.; Xiang, F. Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 2016, 5, 920. [Google Scholar] [CrossRef]
- Li, W.; Tian, Z.; Yu, D. WRKY13 acts in stem development in Arabidopsis thaliana. Plant Sci. 2015, 236, 205–213. [Google Scholar] [CrossRef]
- Wei, L.; Wang, H.; Yu, D. The Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant 2016, 9, 1492–1503. [Google Scholar]
- Jun, X.; Hongtao, C.; Xianghua, L.; Jinghua, X.; Caiguo, X.; Shiping, W. Rice WRKY13 Regulates Cross Talk between Abiotic and Biotic Stress Signaling Pathways by Selective Binding to Different cis-Elements. Plant Phys. 2013, 163, 1868–1882. [Google Scholar]
- Cui, X.Y.; Du, Y.T.; Fu, J.D.; Yu, T.F.; Wang, C.T.; Chen, M.; Chen, J.; Ma, Y.Z.; Xu, Z.S. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 2018, 18, 93. [Google Scholar] [CrossRef]
- Radkova, M.; Vítámvás, P.; Sasaki, K.; Imai, R. Development- and cold-regulated accumulation of cold shock domain proteins in wheat. Plant Physiol. Bioch. 2014, 77, 44–48. [Google Scholar] [CrossRef]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 1997, 275, 80–83. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. Cell Mol. Biol. 2010, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Forno, D.A.; Cock, J.H. Laboratory Manual for Physiological Studies of Rice; CABI: Wallingford, UK, 1971. [Google Scholar]



| Name | ID | Transcript Name | Location |
|---|---|---|---|
| TaWRKY28 | 31740471 | Traes_1BL_9AFA4B870.1 | ta_iwgsc_1bl_v1_3809885:1110..4753 reverse |
| TaWRKY15 | 31742772 | Traes_5BL_E294922A9.2 | ta_iwgsc_5bl_v1_10867378:4708..6402 forward |
| TaWRKY6 | 31744736 | Traes_3B_CDA5ADD75.1 | ta_iwgsc_3b_v1_10758590:296..2989 forward |
| TaWRKY62 | 31745499 | Traes_5DL_C93641E43.1 | ta_iwgsc_5dl_v1_4576731:2459..4427 forward |
| TaWRKY44 | 31746115 | Traes_4AL_2EEECCC4B.1 | ta_iwgsc_4al_v2_7093101:3374..6818 forward |
| TaWRKY74 | 31747511 | Traes_5DL_5C93510D5.1 | ta_iwgsc_5dl_v1_4502975:3368..7661 reverse |
| TaWRKY80 | 31748920 | Traes_6AS_DA75BB1FD.1 | ta_iwgsc_6as_v1_4428654:1..1588 reverse |
| TaWRKY53 | 31752041 | Traes_2DL_F600B5FDF.1 | ta_iwgsc_2dl_v1_9719154:1..728 forward |
| TaWRKY22 | 31752743 | Traes_5AL_6FDB440FB.1 | ta_iwgsc_5al_v1_2705439:4..838 forward |
| TaWRKY45 | 31765470 | Traes_7AL_48C81DE03.1 | ta_iwgsc_7al_v1_4556343:539..3297 forward |
| TaWRKY4 | 31765472 | Traes_7AL_48C81DE031.1 | ta_iwgsc_7al_v1_4556343:3718..6476 reverse |
| TaWRKY35 | 31766778 | Traes_6BL_EEAA2A7E3.1 | ta_iwgsc_6bl_v1_4221964:3..2595 forward |
| TaWRKY79 | 31767242 | Traes_7DL_B09854286.1 | ta_iwgsc_7dl_v1_3393496:18..940 reverse |
| TaWRKY46 | 31768080 | Traes_4DS_FE38A59D0.1 | ta_iwgsc_4ds_v1_2280139:4533..7422 reverse |
| TaWRKY57 | 31782323 | Traes_3B_41047D5E6.2 | ta_iwgsc_3b_v1_10527462:3896..6406 reverse |
| TaWRKY33 | 31785825 | Traes_6DS_8F684013D.1 | ta_iwgsc_6ds_v1_1013038:1..419 reverse |
| TaWRKY12 | 31787421 | Traes_6AL_BA4636569.1 | ta_iwgsc_6al_v1_5754118:409..4140 reverse |
| TaWRKY24 | 31792629 | Traes_4AL_C2A825B6D.1 | ta_iwgsc_4al_v2_3841042:1..253 reverse |
| TaWRKY63 | 31793891 | Traes_3DL_7456F61A3.1 | ta_iwgsc_3dl_v1_5877113:2..2892 reverse |
| TaWRKY68 | 31798439 | Traes_2AL_15A7BB684.1 | ta_iwgsc_2al_v1_6374918:10015..11505 forward |
| TaWRKY50 | 31799212 | Traes_3B_F45FCFE62.1 | ta_iwgsc_3b_v1_10625585:4077..6054 forward |
| TaWRKY58 | 31811544 | Traes_5BL_D3C383CF5.1 | ta_iwgsc_5bl_v1_10787947:2038..3881 forward |
| TaWRKY72 | 31818595 | Traes_2BS_F3097F116.1 | ta_iwgsc_2bs_v1_5195103:6587..11319 forward |
| TaWRKY8 | 31823877 | Traes_5DL_2553A6C33.1 | ta_iwgsc_5dl_v1_4566006:8..1007 forward |
| TaWRKY34 | 31829399 | Traes_2AL_409AB7647.1 | ta_iwgsc_2al_v1_6334600:3412..8916 reverse |
| TaWRKY9 | 31836810 | Traes_2DS_F6FBC974C.2 | ta_iwgsc_2ds_v1_5331381:733..3264 reverse |
| TaWRKY52 | 31851405 | Traes_3AL_AB2BAE660.1 | ta_iwgsc_3al_v1_4270257:1..887 reverse |
| TaWRKY3 | 31853252 | Traes_2DL_4F9F8F1F0.1 | ta_iwgsc_2dl_v1_9906833:634..5055 reverse |
| TaWRKY51 | 31854913 | Traes_2AL_434E9F101.1 | ta_iwgsc_2al_v1_6367445:3985..5752 reverse |
| TaWRKY27 | 31865868 | Traes_3B_990298FF5.1 | ta_iwgsc_3b_v1_10750391:1..2331 reverse |
| TaWRKY70 | 31871499 | Traes_1DL_DFE1721E0.1 | ta_iwgsc_1dl_v1_2268423:4679..7857 forward |
| TaWRKY41 | 31872073 | Traes_2DS_AD8820C42.1 | ta_iwgsc_2ds_v1_5376167:6..1016 reverse |
| TaWRKY14 | 31872762 | Traes_5BL_B9DD3E76F.1 | ta_iwgsc_5bl_v1_10924584:9637..13927 forward |
| TaWRKY17 | 31875786 | Traes_5BL_8688F70C9.1 | ta_iwgsc_5bl_v1_10840877:2232..3827 reverse |
| TaWRKY56 | 31876237 | Traes_3AL_DED8A29EC.1 | ta_iwgsc_3al_v1_382150:704..961 reverse |
| TaWRKY78 | 31876678 | Traes_2BS_D435A8999.1 | ta_iwgsc_2bs_v1_5214231:8279..15893 reverse |
| TaWRKY32 | 31888413 | Traes_4AS_70DF607CC.1 | ta_iwgsc_4as_v2_352920:1884..3594 forward |
| TaWRKY16 | 31891223 | Traes_4AL_98B1C762B.2 | ta_iwgsc_4al_v2_7173949:3935..6881 forward |
| TaWRKY48 | 31892659 | Traes_3B_B8BF316B8.2 | ta_iwgsc_3b_v1_10433739:23..1890 reverse |
| TaWRKY71 | 31894510 | Traes_1DL_46428511F.1 | ta_iwgsc_1dl_v1_2235906:1905..4608 forward |
| TaWRKY38 | 31895081 | Traes_3B_D6F86ABC3.2 | ta_iwgsc_3b_v1_10762199:7310..8822 forward |
| TaWRKY55 | 31916438 | Traes_6AS_68775100B.1 | ta_iwgsc_6as_v1_4413209:7948..9254 forward |
| TaWRKY76 | 31917474 | Traes_5DL_32D78D06A.1 | ta_iwgsc_5dl_v1_4501324:900..1439 reverse |
| TaWRKY19 | 31924920 | Traes_2BS_380EC4D1E.1 | ta_iwgsc_2bs_v1_5227909:9257..13033 reverse |
| TaWRKY29 | 31938855 | Traes_1AL_4E924201A.1 | ta_iwgsc_1al_v2_3969710:4988..6878 reverse |
| TaWRKY10 | 31942345 | Traes_2DL_362A1F535.1 | ta_iwgsc_2dl_v1_9707610:58..446 forward |
| TaWRKY36 | 31942939 | Traes_3AL_140B829CB.2 | ta_iwgsc_3al_v1_4308486:3673..5708 reverse |
| TaWRKY2 | 31951792 | Traes_5BL_17A712C94.1 | ta_iwgsc_5bl_v1_10916210:5033..9621 forward |
| TaWRKY13 | 31962353 | Traes_2AS_6269D889E.1 | ta_iwgsc_2as_v1_5205891:13214..14843 reverse |
| TaWRKY1 | 31966248 | Traes_5BL_AEF9FE805.1 | ta_iwgsc_5bl_v1_10827243:3081..6424 reverse |
| TaWRKY49 | 31968771 | Traes_3DL_2551BF2C1.1 | ta_iwgsc_3dl_v1_6811598:1..1035 reverse |
| TaWRKY23 | 31977027 | Traes_3AL_4769A72F1.1 | ta_iwgsc_3al_v1_805190:2..774 reverse |
| TaWRKY75 | 31987126 | Traes_1AL_0404BC790.1 | ta_iwgsc_1al_v2_3912777:3..1909 forward |
| TaWRKY5 | 31988149 | Traes_5AL_7164FEAC3.1 | ta_iwgsc_5al_v1_2204788:3..342 forward |
| TaWRKY64 | 32002393 | Traes_4AS_0DA136E0E.1 | ta_iwgsc_4as_v2_5962726:2807..4440 forward |
| TaWRKY90 | 32002429 | Traes_3AL_1B73D2C12.1 | ta_iwgsc_3al_v1_4248344:659..1678 forward |
| TaWRKY61 | 32024774 | Traes_5BS_C46781248.1 | ta_iwgsc_5bs_v1_2248873:15934..19692 reverse |
| Cis-Element | Target Sequences | Number | Function |
|---|---|---|---|
| W-BOX | TTGAC/TGACT TGACY/CTCAY | 27 | Drought, high salt responsive elements |
| MYB | GGATA/WAACCA/TAACARA/ TAACAAA/CCWACC/GNGTTR | 20 | Drought, high salt responsive elements |
| LTR | CCGAC/CCGAAA | 7 | Low-temperature, salt responsive elements |
| ABRE | ACGTGKC | 5 | ABA-responsive elements |
| TATA-BOX | TATATAA | 6 | Drought, cold, high salt responsive elements |
| GTI | CAAAAA | 3 | Salt responsive elements |
| GATA-BOX | GATA | 22 | Light, gibberellin responsive elements |
| WRKY | TAGA | 20 | Light, salicylic acid responsive elements |
| HSP70A | SCGAYNR(N)15HD | 7 | High temperature responsive elements |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, S.; Zheng, W.-J.; Liu, B.-H.; Zheng, J.-C.; Dong, F.-S.; Liu, Z.-F.; Wen, Z.-Y.; Yang, F.; Wang, H.-B.; Xu, Z.-S.; et al. Characterizing the Role of TaWRKY13 in Salt Tolerance. Int. J. Mol. Sci. 2019, 20, 5712. https://doi.org/10.3390/ijms20225712
Zhou S, Zheng W-J, Liu B-H, Zheng J-C, Dong F-S, Liu Z-F, Wen Z-Y, Yang F, Wang H-B, Xu Z-S, et al. Characterizing the Role of TaWRKY13 in Salt Tolerance. International Journal of Molecular Sciences. 2019; 20(22):5712. https://doi.org/10.3390/ijms20225712
Chicago/Turabian StyleZhou, Shuo, Wei-Jun Zheng, Bao-Hua Liu, Jia-Cheng Zheng, Fu-Shuang Dong, Zhi-Fang Liu, Zhi-Yu Wen, Fan Yang, Hai-Bo Wang, Zhao-Shi Xu, and et al. 2019. "Characterizing the Role of TaWRKY13 in Salt Tolerance" International Journal of Molecular Sciences 20, no. 22: 5712. https://doi.org/10.3390/ijms20225712
APA StyleZhou, S., Zheng, W.-J., Liu, B.-H., Zheng, J.-C., Dong, F.-S., Liu, Z.-F., Wen, Z.-Y., Yang, F., Wang, H.-B., Xu, Z.-S., Zhao, H., & Liu, Y.-W. (2019). Characterizing the Role of TaWRKY13 in Salt Tolerance. International Journal of Molecular Sciences, 20(22), 5712. https://doi.org/10.3390/ijms20225712







