Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells
Abstract
1. Introduction
2. Results
2.1. Expression of PTX3 during Osteo/Odontogenic Differentiation of HDPSCs
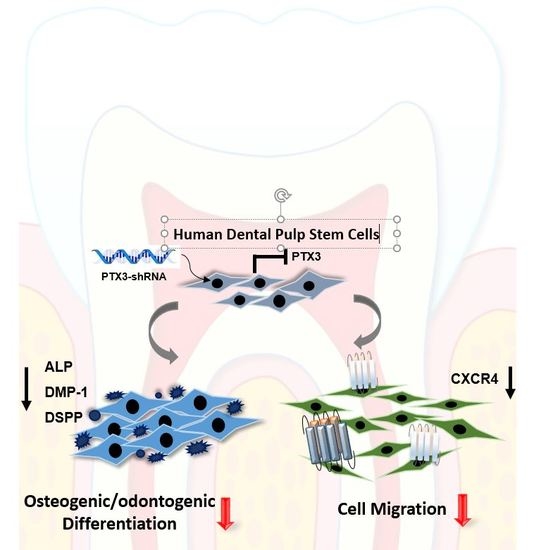
2.2. Inhibition of PTX3 Impairs Osteo/Odontogenic Differentiation of HDPSCs
2.3. Knockdown of PTX3 Inhibits Migration of HDPSCs
2.4. Knockdown of PTX3 Regulates SDF-1/CXCR4 Axis in HDPSCs
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. shRNA Lentiviral Particle Transduction
4.4. Cell Proliferation Assay
4.5. Alkaline Phosphatase (ALP) Staining
4.6. Real-Time PCR Analysis
4.7. Western Immunoblot Analysis
4.8. Enzyme-Linked Immunosorbent Assays (ELISA)
4.9. Transwell Chemotactic Migration Assay
4.10. Scratch Wound Migration Assay
4.11. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Cooper, P.R.; Holder, M.J.; Smith, A.J. Inflammation and Regeneration in the Dentin-Pulp Complex: A Double-Edged Sword. J. Endod. 2014, 40, S46–S51. [Google Scholar] [CrossRef]
- Masthan, K.M.; Sankari, S.L.; Babu, N.A.; Gopalakrishnan, T. Mystery Inside the Tooth: The Dental Pulp Stem Cells. J. Clin. Diagn. Res. 2013, 7, 945–947. [Google Scholar] [CrossRef] [PubMed]
- Fawzy El-Sayed, K.M.; Elsalawy, R.; Ibrahim, N.; Gadalla, M.; Albargasy, H.; Zahra, N.; Mokhtar, S.; Elnahhas, N.; Elkaliouby, Y.; Doerfer, C. The Dental Pulp stem/progenitor Cells-Mediated Inflammatory-Regenerative Axis. Tissue Eng. Part B. Rev. 2019, 25, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Vilahur, G.; Badimon, L. Biological Actions of Pentraxins. Vascul Pharmacol. 2015, 73, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Garlanda, C.; Bottazzi, B.; Peri, G.; Doni, A.; Martinez de la Torre, Y.; Latini, R. The Long Pentraxin PTX3 in Vascular Pathology. Vascul Pharmacol. 2006, 45, 326–330. [Google Scholar] [CrossRef]
- Garlanda, C.; Bottazzi, B.; Magrini, E.; Inforzato, A.; Mantovani, A. PTX3, a Humoral Pattern Recognition Molecule, in Innate Immunity, Tissue Repair, and Cancer. Physiol. Rev. 2018, 98, 623–639. [Google Scholar] [CrossRef]
- Daigo, K.; Mantovani, A.; Bottazzi, B. The Yin-Yang of Long Pentraxin PTX3 in Inflammation and Immunity. Immunol. Lett. 2014, 161, 38–43. [Google Scholar] [CrossRef]
- Kim, Y.; Park, J.S.; Park, H.J.; Kim, M.K.; Kim, Y.I.; Bae, S.K.; Kim, H.J.; Jeong, C.H.; Bae, M.K. Pentraxin 3 Modulates the Inflammatory Response in Human Dental Pulp Cells. J. Endod. 2018, 44, 1826–1831. [Google Scholar] [CrossRef]
- Gong, Q.M.; Quan, J.J.; Jiang, H.W.; Ling, J.Q. Regulation of the Stromal Cell-Derived Factor-1alpha-CXCR4 Axis in Human Dental Pulp Cells. J. Endod. 2010, 36, 1499–1503. [Google Scholar] [CrossRef]
- Jiang, H.W.; Ling, J.Q.; Gong, Q.M. The Expression of Stromal Cell-Derived Factor 1 (SDF-1) in Inflamed Human Dental Pulp. J. Endod. 2008, 34, 1351–1354. [Google Scholar] [CrossRef]
- Suzuki, T.; Lee, C.H.; Chen, M.; Zhao, W.; Fu, S.Y.; Qi, J.J.; Chotkowski, G.; Eisig, S.B.; Wong, A.; Mao, J.J. Induced Migration of Dental Pulp Stem Cells for in Vivo Pulp Regeneration. J. Dent. Res. 2011, 90, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, X.; Ma, L.; Jin, L.; Zhang, W.; Xiao, M.; Yu, Q. SDF-1/CXCR4 Axis Induces Human Dental Pulp Stem Cell Migration through FAK/PI3K/Akt and GSK3beta/beta-Catenin Pathways. Sci. Rep. 2017, 7, 40161. [Google Scholar] [CrossRef] [PubMed]
- Fornai, F.; Carrizzo, A.; Forte, M.; Ambrosio, M.; Damato, A.; Ferrucci, M.; Biagioni, F.; Busceti, C.; Puca, A.A.; Vecchione, C. The Inflammatory Protein Pentraxin 3 in Cardiovascular Disease. Immun. Ageing 2016, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Kunes, P.; Holubcova, Z.; Kolackova, M.; Krejsek, J. Pentraxin 3(PTX 3): An Endogenous Modulator of the Inflammatory Response. Mediators Inflamm. 2012, 2012, 920517. [Google Scholar] [CrossRef]
- Magrini, E.; Mantovani, A.; Garlanda, C. The Dual Complexity of PTX3 in Health and Disease: A Balancing Act? Trends Mol. Med. 2016, 22, 497–510. [Google Scholar] [CrossRef]
- Real, J.M.; Spilborghs, G.M.; Morato-Marques, M.; de Moura, R.P.; Negri, E.M.; Camargo, A.A.; Deheinzelin, D.; Dias, A.A. Pentraxin 3 Accelerates Lung Injury in High Tidal Volume Ventilation in Mice. Mol. Immunol. 2012, 51, 82–90. [Google Scholar] [CrossRef]
- Carrizzo, A.; Lenzi, P.; Procaccini, C.; Damato, A.; Biagioni, F.; Ambrosio, M.; Amodio, G.; Remondelli, P.; Del Giudice, C.; Izzo, R.; et al. Pentraxin 3 Induces Vascular Endothelial Dysfunction through a P-selectin/Matrix Metalloproteinase-1 Pathway. Circulation 2015, 131, 1495–1505. [Google Scholar] [CrossRef]
- Souza, D.G.; Amaral, F.A.; Fagundes, C.T.; Coelho, F.M.; Arantes, R.M.; Sousa, L.P.; Matzuk, M.M.; Garlanda, C.; Mantovani, A.; Dias, A.A.; et al. The Long Pentraxin PTX3 is Crucial for Tissue Inflammation After Intestinal Ischemia and Reperfusion in Mice. Am. J. Pathol. 2009, 174, 1309–1318. [Google Scholar] [CrossRef]
- Dias, A.A.; Goodman, A.R.; Dos Santos, J.L.; Gomes, R.N.; Altmeyer, A.; Bozza, P.T.; Horta, M.F.; Vilcek, J.; Reis, L.F. TSG-14 Transgenic Mice have Improved Survival to Endotoxemia and to CLP-Induced Sepsis. J. Leukoc. Biol. 2001, 69, 928–936. [Google Scholar]
- Rodriguez-Grande, B.; Swana, M.; Nguyen, L.; Englezou, P.; Maysami, S.; Allan, S.M.; Rothwell, N.J.; Garlanda, C.; Denes, A.; Pinteaux, E. The Acute-Phase Protein PTX3 is an Essential Mediator of Glial Scar Formation and Resolution of Brain Edema After Ischemic Injury. J. Cereb. Blood Flow Metab. 2014, 34, 480–488. [Google Scholar] [CrossRef]
- Deban, L.; Russo, R.C.; Sironi, M.; Moalli, F.; Scanziani, M.; Zambelli, V.; Cuccovillo, I.; Bastone, A.; Gobbi, M.; Valentino, S.; et al. Regulation of Leukocyte Recruitment by the Long Pentraxin PTX3. Nat. Immunol. 2010, 11, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Cuello, F.; Shankar-Hari, M.; Mayr, U.; Yin, X.; Marshall, M.; Suna, G.; Willeit, P.; Langley, S.R.; Jayawardhana, T.; Zeller, T.; et al. Redox State of Pentraxin 3 as a Novel Biomarker for Resolution of Inflammation and Survival in Sepsis. Mol. Cell. Proteomics 2014, 13, 2545–2557. [Google Scholar] [CrossRef] [PubMed]
- Kizil, C.; Kyritsis, N.; Brand, M. Effects of Inflammation on Stem Cells: Together they Strive? EMBO Rep. 2015, 16, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Ye, L.; Love, R.M.; Farges, J.C.; Yumoto, H. Inflammation of the Dental Pulp. Mediators Inflamm. 2015, 2015, 980196. [Google Scholar] [CrossRef]
- Cooper, P.R.; Takahashi, Y.; Graham, L.W.; Simon, S.; Imazato, S.; Smith, A.J. Inflammation-Regeneration Interplay in the Dentine-Pulp Complex. J. Dent. 2010, 38, 687–697. [Google Scholar] [CrossRef]
- Alongi, D.J.; Yamaza, T.; Song, Y.; Fouad, A.F.; Romberg, E.E.; Shi, S.; Tuan, R.S.; Huang, G.T. Stem/progenitor Cells from Inflamed Human Dental Pulp Retain Tissue Regeneration Potential. Regen. Med. 2010, 5, 617–631. [Google Scholar] [CrossRef]
- Bastrup-Birk, S.; Skjoedt, M.O.; Munthe-Fog, L.; Strom, J.J.; Ma, Y.J.; Garred, P. Pentraxin-3 Serum Levels are Associated with Disease Severity and Mortality in Patients with Systemic Inflammatory Response Syndrome. PLoS ONE 2013, 8, e73119. [Google Scholar] [CrossRef]
- Liu, S.; Qu, X.; Liu, F.; Wang, C. Pentraxin 3 as a Prognostic Biomarker in Patients with Systemic Inflammation or Infection. Mediators Inflamm. 2014, 2014, 421429. [Google Scholar] [CrossRef]
- Kotooka, N.; Inoue, T.; Fujimatsu, D.; Morooka, T.; Hashimoto, S.; Hikichi, Y.; Uchida, T.; Sugiyama, A.; Node, K. Pentraxin3 is a Novel Marker for Stent-Induced Inflammation and Neointimal Thickening. Atherosclerosis 2008, 197, 368–374. [Google Scholar] [CrossRef]
- Stallone, G.; Cormio, L.; Netti, G.S.; Infante, B.; Selvaggio, O.; Fino, G.D.; Ranieri, E.; Bruno, F.; Prattichizzo, C.; Sanguedolce, F.; et al. Pentraxin 3: A Novel Biomarker for Predicting Progression from Prostatic Inflammation to Prostate Cancer. Cancer Res. 2014, 74, 4230–4238. [Google Scholar] [CrossRef]
- Gumus, P.; Nizam, N.; Nalbantsoy, A.; Ozcaka, O.; Buduneli, N. Saliva and Serum Levels of Pentraxin-3 and Interleukin-1beta in Generalized Aggressive or Chronic Periodontitis. J. Periodontol. 2014, 85, e40–e46. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, A.R.; Kathariya, R.; Raghavendra, N.M.; Sharma, A. Levels of Pentraxin-3 in Gingival Crevicular Fluid and Plasma in Periodontal Health and Disease. J. Periodontol. 2011, 82, 734–741. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.; Farges, J.C.; Lacerda-Pinheiro, S.; Six, N.; Jegat, N.; Decup, F.; Septier, D.; Carrouel, F.; Durand, S.; Chaussain-Miller, C.; et al. Inflammatory and Immunological Aspects of Dental Pulp Repair. Pharmacol. Res. 2008, 58, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Tziafas, D.; Smith, A.J.; Lesot, H. Designing New Treatment Strategies in Vital Pulp Therapy. J. Dent. 2000, 28, 77–92. [Google Scholar] [CrossRef]
- Imitola, J.; Raddassi, K.; Park, K.I.; Mueller, F.J.; Nieto, M.; Teng, Y.D.; Frenkel, D.; Li, J.; Sidman, R.L.; Walsh, C.A.; et al. Directed Migration of Neural Stem Cells to Sites of CNS Injury by the Stromal Cell-Derived Factor 1alpha/CXC Chemokine Receptor 4 Pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 18117–18122. [Google Scholar] [CrossRef]
- Jiang, L.; Zhu, Y.Q.; Du, R.; Gu, Y.X.; Xia, L.; Qin, F.; Ritchie, H.H. The Expression and Role of Stromal Cell-Derived Factor-1alpha-CXCR4 Axis in Human Dental Pulp. J. Endod. 2008, 34, 939–944. [Google Scholar] [CrossRef]
- Kim, D.S.; Kim, Y.S.; Bae, W.J.; Lee, H.J.; Chang, S.W.; Kim, W.S.; Kim, E.C. The Role of SDF-1 and CXCR4 on Odontoblastic Differentiation in Human Dental Pulp Cells. Int. Endod. J. 2014, 47, 534–541. [Google Scholar] [CrossRef]
- Jiang, L.; Peng, W.W.; Li, L.F.; Yang, Y.; Zhu, Y.Q. Proliferation and Multilineage Potential of CXCR4-Positive Human Dental Pulp Cells in Vitro. J. Endod. 2012, 38, 642–647. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Park, J.-Y.; Park, H.-J.; Kim, M.-K.; Kim, Y.-I.; Kim, H.J.; Bae, S.-K.; Bae, M.-K. Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells. Int. J. Mol. Sci. 2019, 20, 5778. https://doi.org/10.3390/ijms20225778
Kim Y, Park J-Y, Park H-J, Kim M-K, Kim Y-I, Kim HJ, Bae S-K, Bae M-K. Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells. International Journal of Molecular Sciences. 2019; 20(22):5778. https://doi.org/10.3390/ijms20225778
Chicago/Turabian StyleKim, Yeon, Joo-Yeon Park, Hyun-Joo Park, Mi-Kyoung Kim, Yong-Il Kim, Hyung Joon Kim, Soo-Kyung Bae, and Moon-Kyoung Bae. 2019. "Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells" International Journal of Molecular Sciences 20, no. 22: 5778. https://doi.org/10.3390/ijms20225778
APA StyleKim, Y., Park, J.-Y., Park, H.-J., Kim, M.-K., Kim, Y.-I., Kim, H. J., Bae, S.-K., & Bae, M.-K. (2019). Pentraxin-3 Modulates Osteogenic/Odontogenic Differentiation and Migration of Human Dental Pulp Stem Cells. International Journal of Molecular Sciences, 20(22), 5778. https://doi.org/10.3390/ijms20225778