Carotenoid Cleavage Dioxygenases: Identification, Expression, and Evolutionary Analysis of This Gene Family in Tobacco
Abstract
1. Introduction
2. Results
2.1. Identification and Characterization of CCD
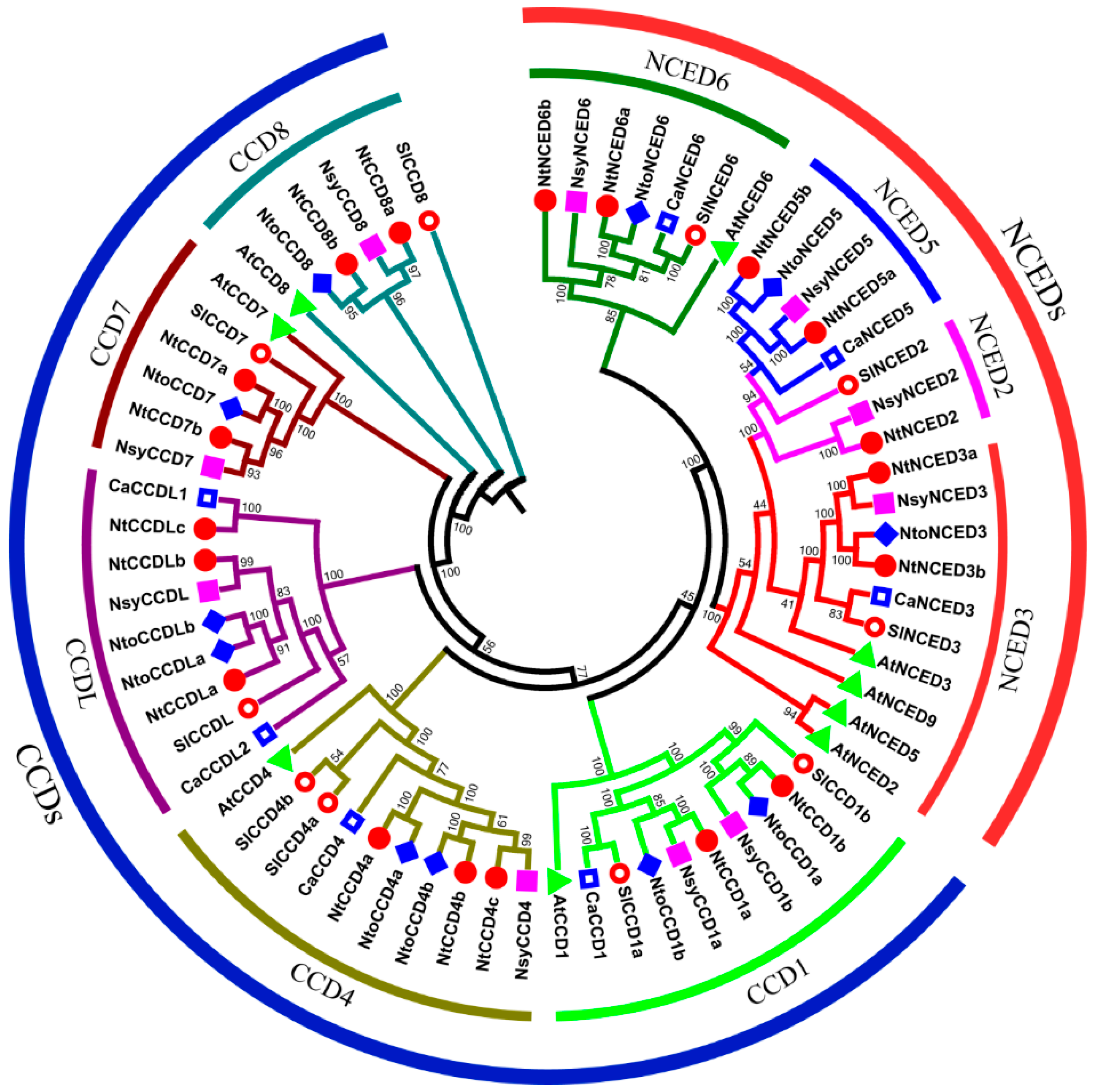
2.2. Phylogenetic Analysis of CCD
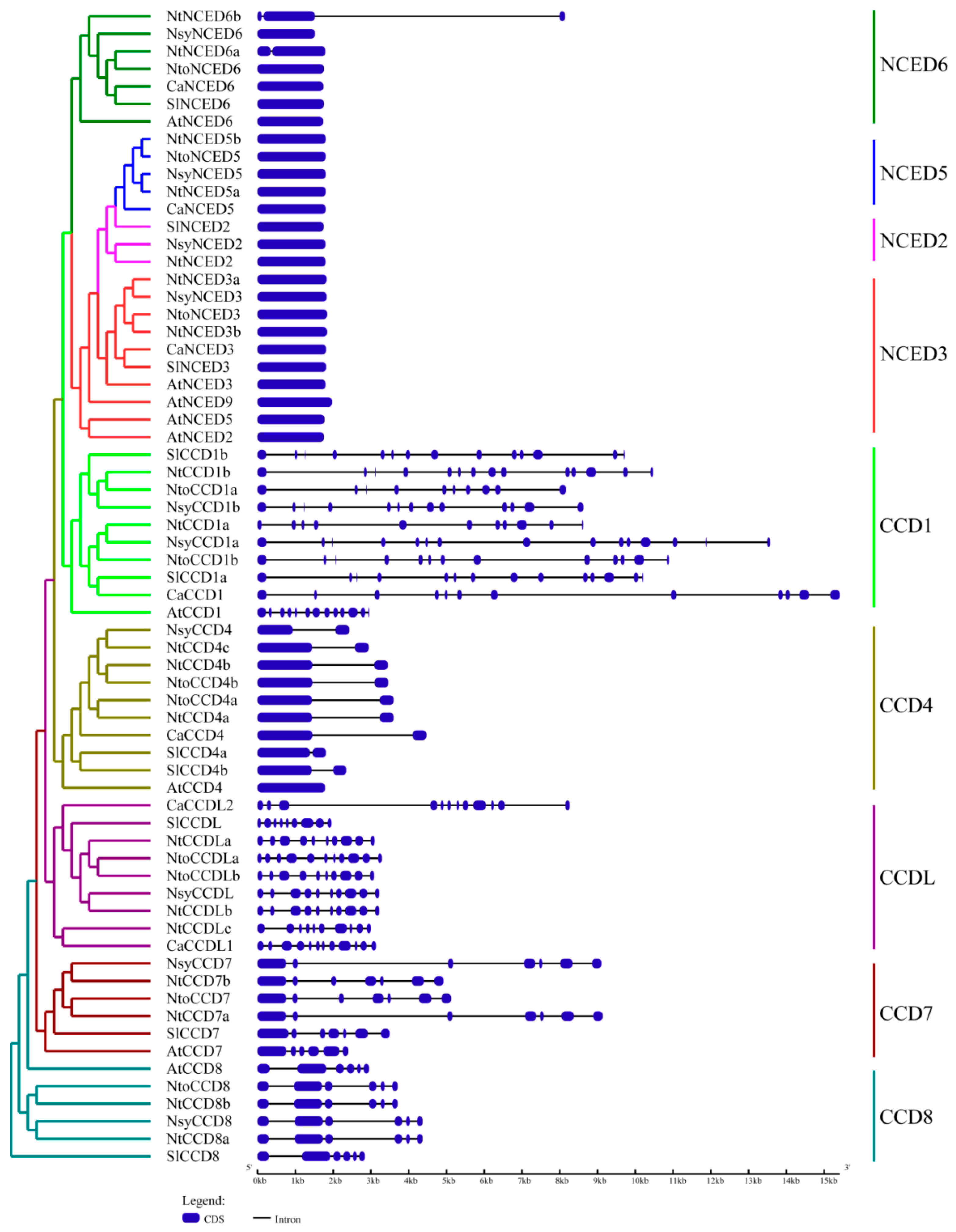
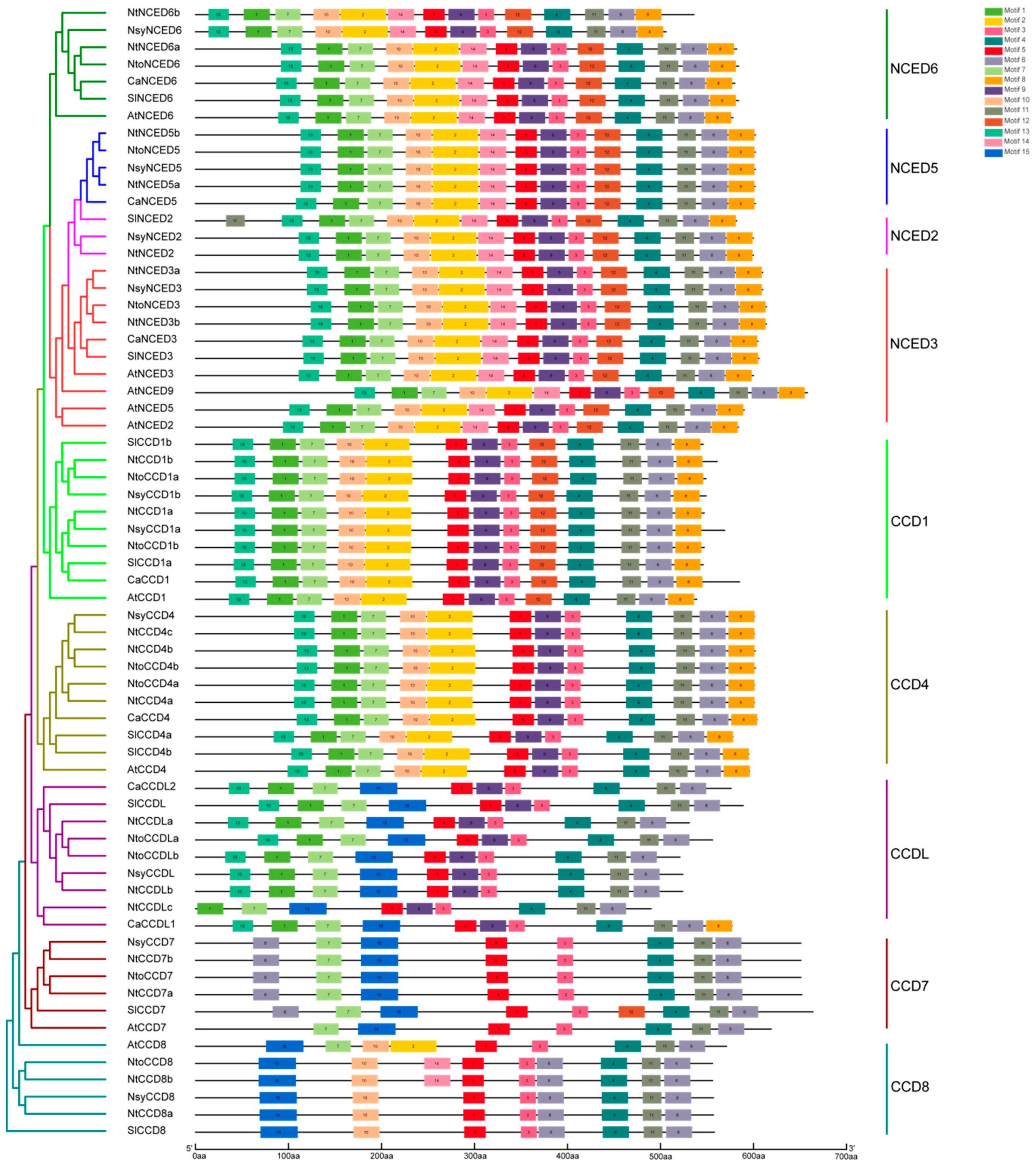
2.3. Gene Structures and Conserved Motifs Analysis of CCDs
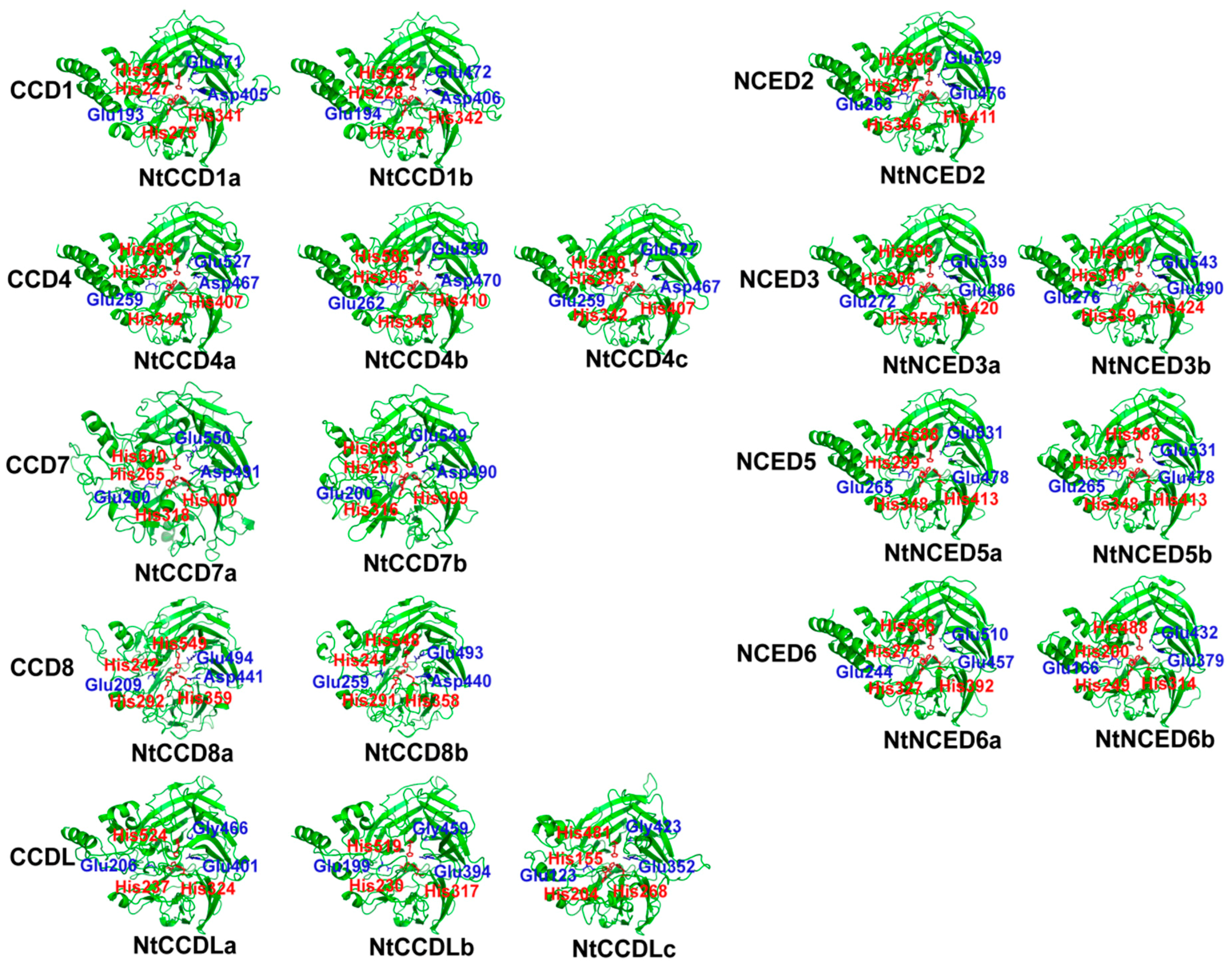
2.4. Predicted Three-Dimensional (3D) Structures of NtCCDs
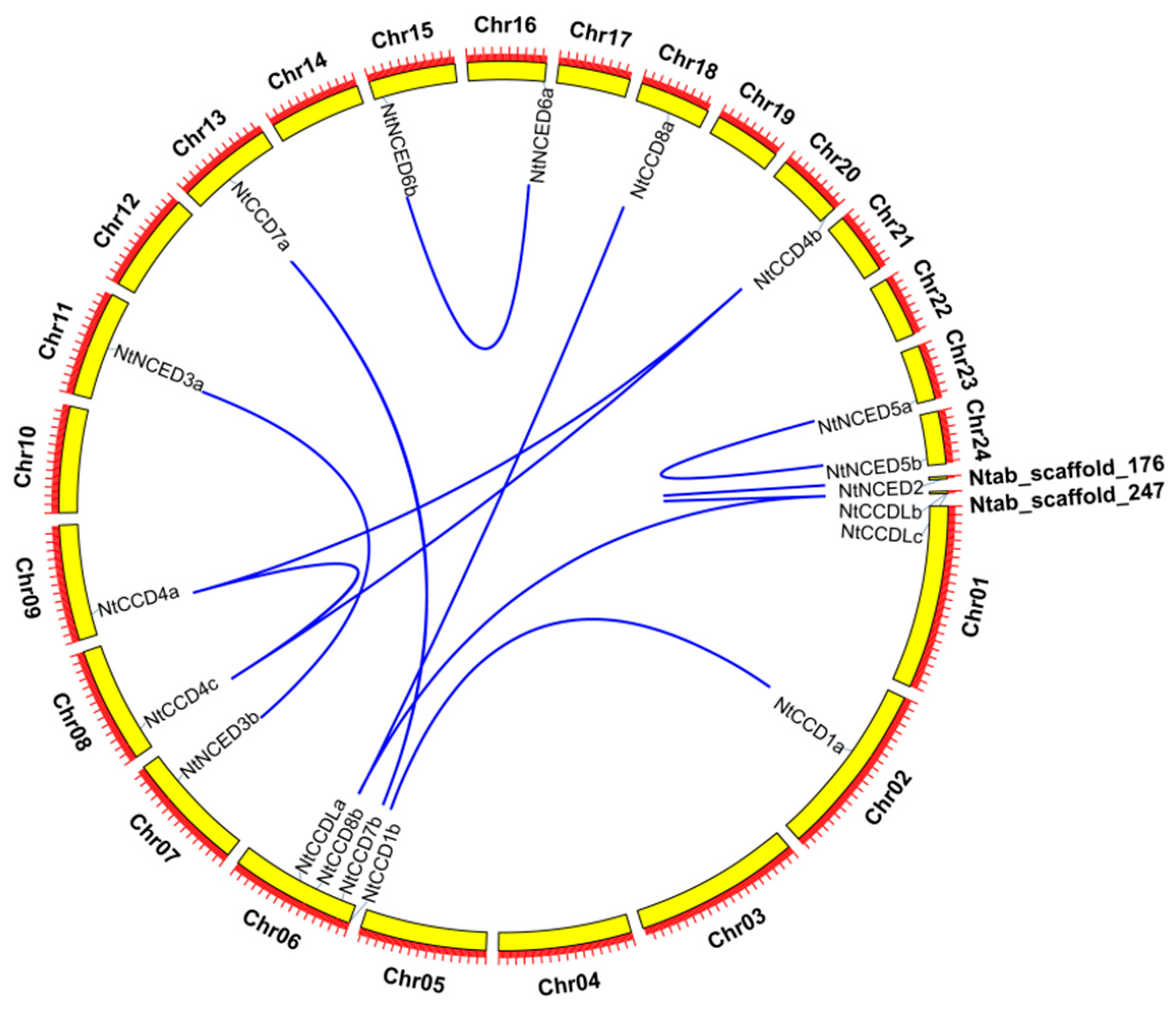
2.5. Chromosomal Locations and Gene Duplication Analysis of NtCCDs
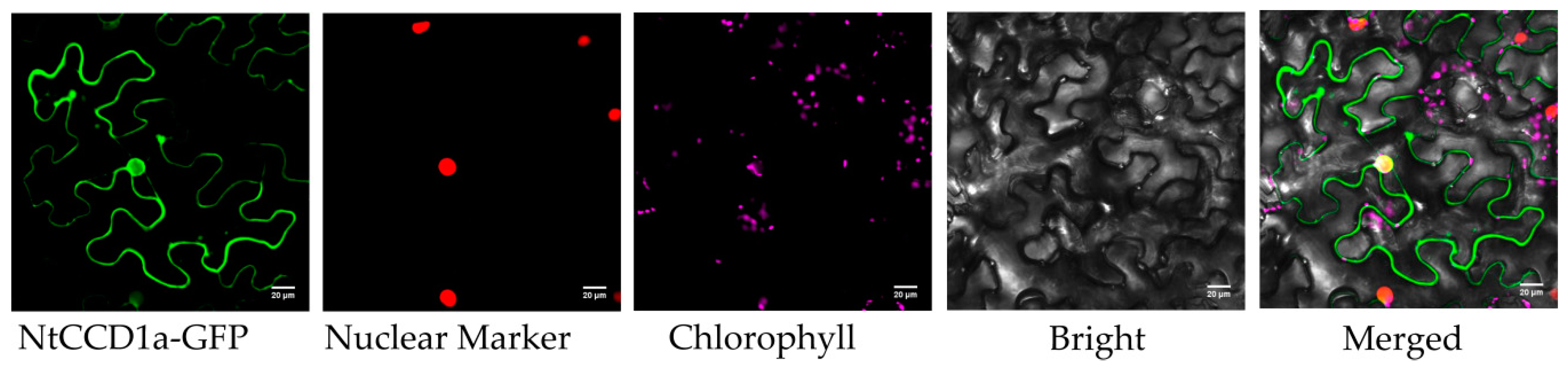
2.6. Subcellular Localization of CCD Proteins
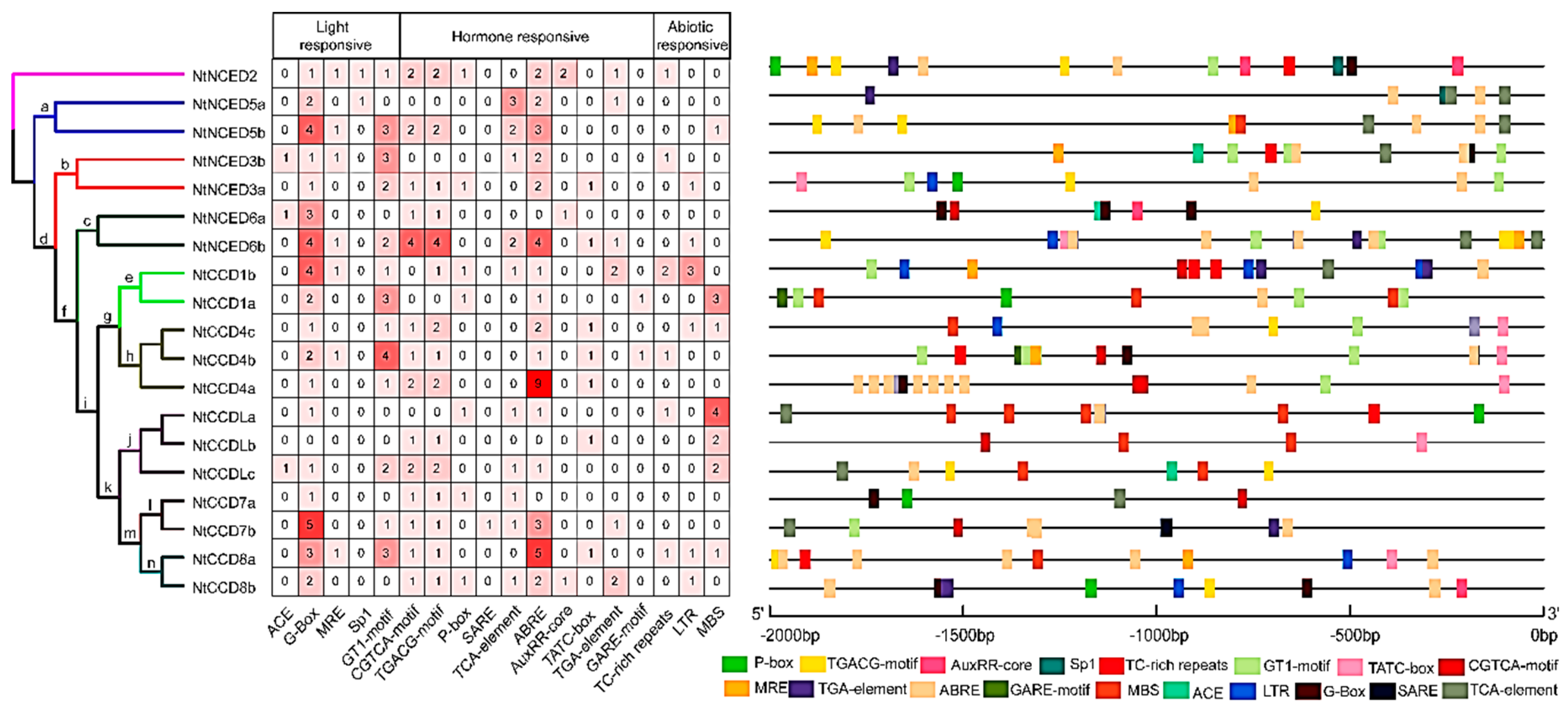
2.7. Cis-acting Elements Analysis of NtCCDs Promoters
2.8. Potential MicroRNA Target Sites of NtCCDs
2.9. Molecular Evolution of NtCCDs
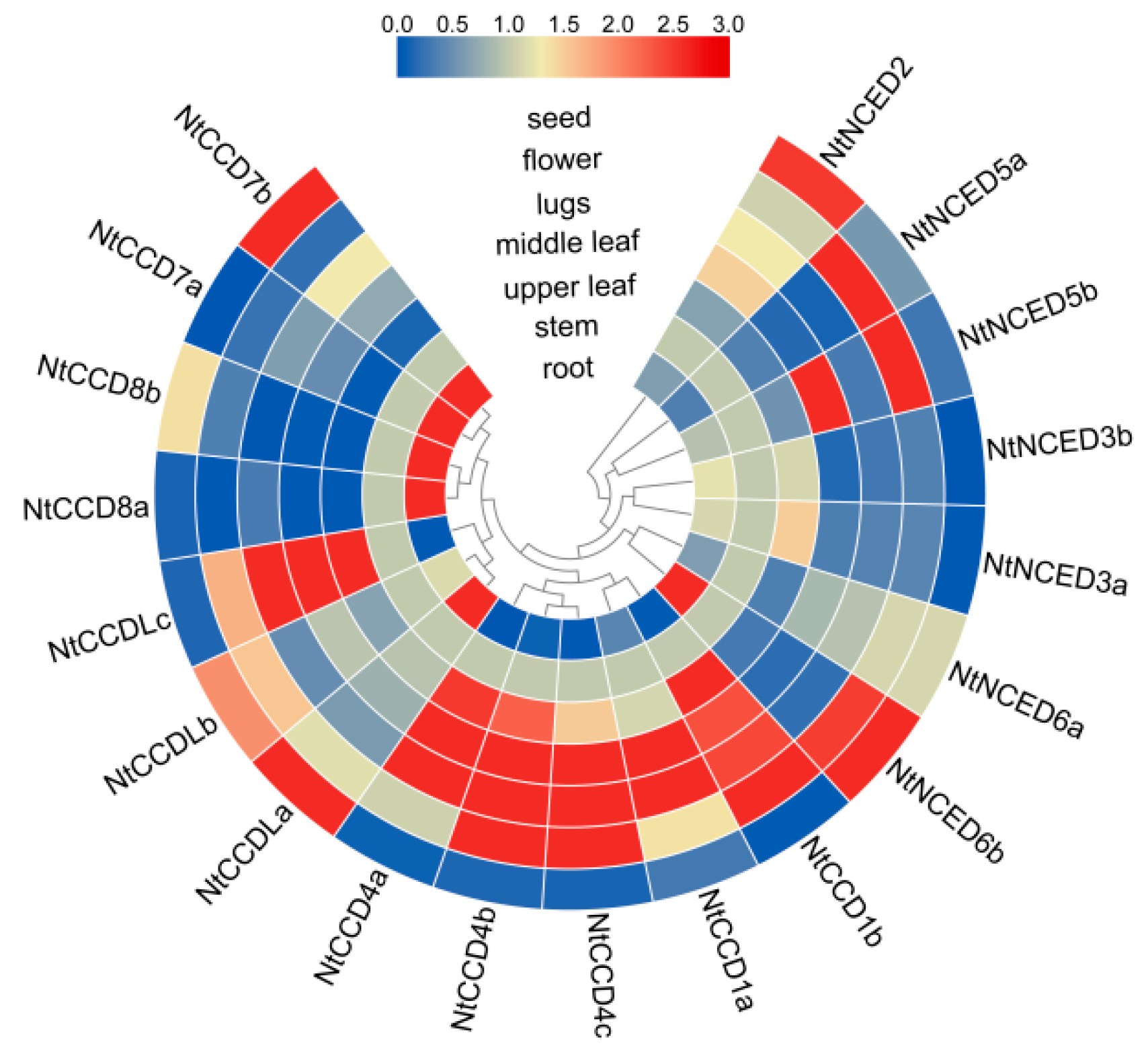
2.10. Expression Patterns of NtCCDs in Different Tissues
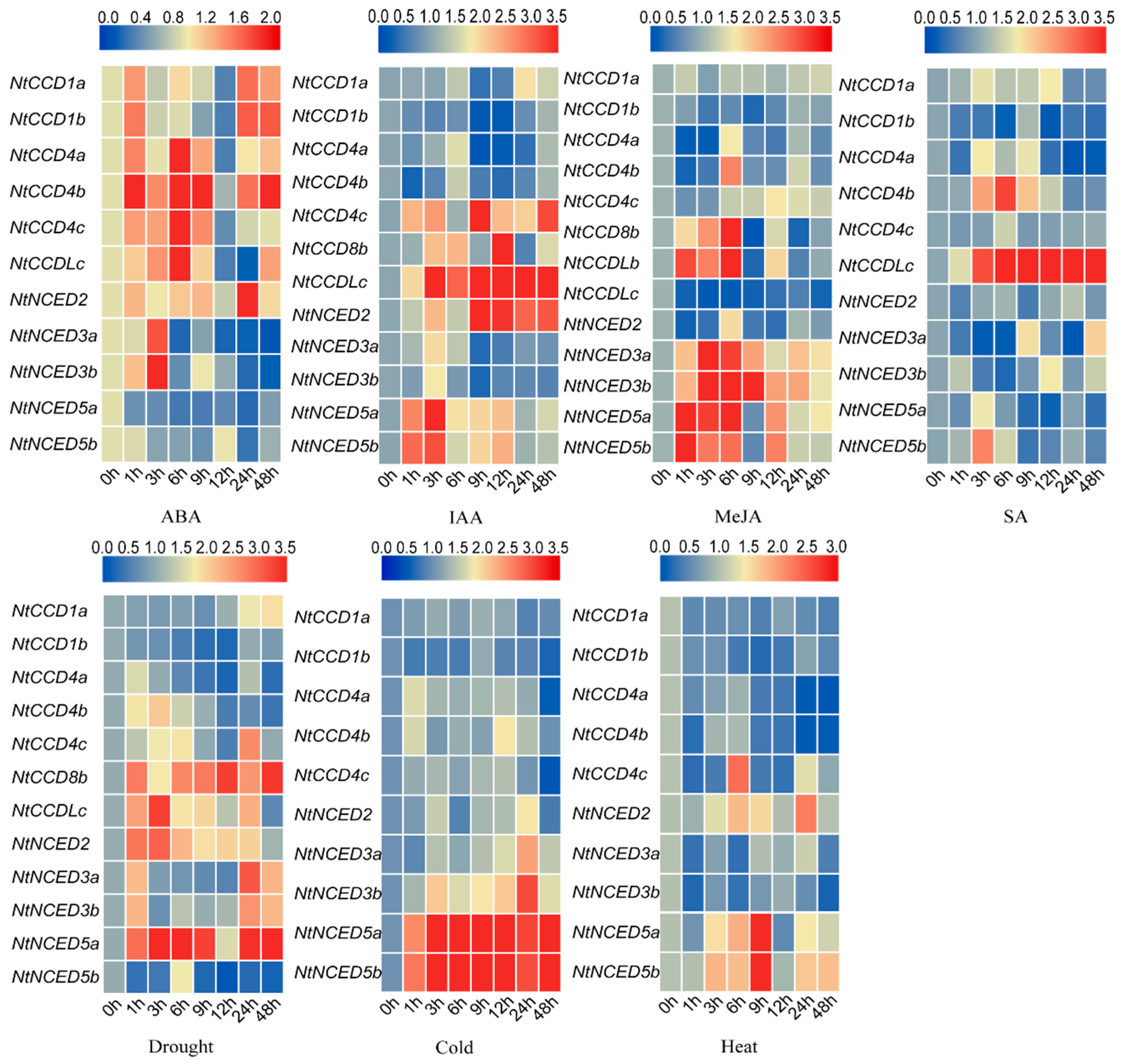
2.11. Gene Expression Analysis of Responses to Hormone Treatments and Abiotic Stresses
3. Discussion
3.1. Basic Characteristics of NtCCD Genes Family
3.2. Molecular Evolution of NtCCDs Genes in Tobacco
3.3. Regulation of NtCCD Genes in Tobacco
4. Materials and Methods
4.1. Identification and Characterization of CCD
4.2. Phylogenetic Analysis of CCD Proteins and Gene Nomenclature
4.3. Gene Structures and Conserved-Motif Prediction
4.4. Three-Dimensional (3D) Structure Modeling of CCD Family Proteins
4.5. Chromosomal Locations and Gene Duplication Analysis of CCD Genes in Tobacco
4.6. Subcellular Localization
4.7. MiRNA Target Sites and Cis-Acting Regulatory Elements Prediction
4.8. Molecular Evolution of CCD Genes in Tobacco
4.9. Plant Materials and Treatments
4.10. RNA Extraction, Reverse Transcription, and Quantitative Reverse-Transcription PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CCD | Carotenoid cleavage dioxygenase |
| NCED | 9-Cis-epoxycarotenoid dioxygenase |
| ABA | Abscisic acid |
| SL | Strigolactone |
| IAA | Indole-3-acetic acid |
| SA | Salicylic acid |
| Mw | Molecular weight |
| pI | Isoelectric point |
| ACE | Cis-acting element involved in light responsiveness |
| G-Box | Cis-acting regulatory element involved in light responsiveness |
| MRE | MYB binding site involved in light responsiveness |
| Sp1 | Light responsive element |
| GT1-motif | Light responsive element |
| CGTCA-motif | Cis-acting regulatory element involved in the MeJA-responsiveness |
| TGACG-motif | Cis-acting regulatory element involved in the MeJA-responsiveness |
| P-box | Gibberellin-responsive element |
| SARE | Cis-acting element involved in salicylic acid responsiveness |
| TCA-element | Cis-acting element involved in salicylic acid responsiveness |
| ABRE | Cis-acting element involved in the abscisic acid responsiveness |
| AuxRR-core | Cis-acting regulatory element involved in auxin responsiveness |
| TATC-box | Cis-acting element involved in gibberellin-responsiveness |
| TGA-element | Auxin-responsive element |
| GARE-motif | Gibberellin-responsive element |
| TC-rich repeats | Cis-acting element involved in defense and stress responsiveness |
| LTR | Cis-acting element involved in low-temperature responsiveness |
| MBS | MYB binding site involved in drought-inducibility |
References
- Ahrazem, O.; Gomez-Gomez, L.; Rodrigo, M.J.; Avalos, J.; Limon, M.C. Carotenoid Cleavage Oxygenases from Microbes and Photosynthetic Organisms: Features and Functions. Int. J. Mol. Sci. 2016, 17, 1781. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 2009, 26, 351–358. [Google Scholar] [CrossRef]
- Hou, X.; Rivers, J.; Leon, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and Function of Apocarotenoid Signals in Plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Floss, D.S.; Strack, D. Apocarotenoids: Hormones, mycorrhizal metabolites and aroma volatiles. Planta 2010, 232, 1–17. [Google Scholar] [CrossRef] [PubMed]
- McQuinn, R.P.; Giovannoni, J.J.; Pogson, B.J. More than meets the eye: From carotenoid biosynthesis, to new insights into apocarotenoid signaling. Curr. Opin. Plant Biol. 2015, 27, 172–179. [Google Scholar] [CrossRef]
- Ma, G.; Zhang, L.; Matsuta, A.; Matsutani, K.; Yamawaki, K.; Yahata, M.; Wahyudi, A.; Motohashi, R.; Kato, M. Enzymatic formation of β-citraurin from β-cryptoxanthin and Zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 2013, 163, 682–695. [Google Scholar] [CrossRef]
- Huang, F.C.; Molnar, P.; Schwab, W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 2009, 60, 3011–3022. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific Oxidative Cleavage of Carotenoids by VP14 of Maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.J.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Wang, R.K.; Wang, C.E.; Fei, Y.Y.; Gai, J.Y.; Zhao, T.J. Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol. Biol. Rep. 2013, 40, 4737–4745. [Google Scholar] [CrossRef]
- Wei, Y.P.; Wan, H.J.; Wu, Z.M.; Wang, R.Q.; Ruan, M.Y.; Ye, Q.J.; Li, Z.M.; Zhou, G.Z.; Yao, Z.P.; Yang, Y.J. A Comprehensive Analysis of Carotenoid Cleavage Dioxygenases Genes in Solanum Lycopersicum. Plant Mol. Biol. Rep. 2016, 34, 512–523. [Google Scholar] [CrossRef]
- Wang, Y.; Ding, G.J.; Gu, T.T.; Ding, J.; Li, Y. Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol. Genet. Genomics 2017, 292, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Zuo, X.Y.; Shao, H.X.; Fan, S.; Ma, J.J.; Zhang, D.; Zhao, C.P.; Yan, X.Y.; Liu, X.J.; Han, M.Y. Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol. Biochem. 2018, 123, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Berman, J.; Capell, T.; Christou, P.; Zhu, C.; Gomez-Gomez, L. The carotenoid cleavage dioxygenase CCD2 catalysing the synthesis of crocetin in spring crocuses and saffron is a plastidial enzyme. New Phytol. 2016, 209, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Ahrazem, O.; Rubio-Moraga, A.; Argandona-Picazo, J.; Castillo, R.; Gomez-Gomez, L. Intron retention and rhythmic diel pattern regulation of carotenoid cleavage dioxygenase 2 during crocetin biosynthesis in saffron. Plant Mol. Biol. 2016, 91, 355–374. [Google Scholar] [CrossRef]
- Sun, Z.; Hans, J.; Walter, M.H.; Matusova, R.; Beekwilder, J.; Verstappen, F.W.; Ming, Z.; van Echtelt, E.; Strack, D.; Bisseling, T.; et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 2008, 228, 789–801. [Google Scholar] [CrossRef]
- Yu, J.; Ai, G.; Shen, D.Y.; Chai, C.Y.; Jia, Y.L.; Liu, W.J.; Dou, D.L. Bioinformatical analysis and prediction of Nicotiana benthamiana bHLH transcription factors in Phytophthora parasitica resistance. Genomics 2019, 111, 473–482. [Google Scholar] [CrossRef]
- Ilg, A.; Beyer, P.; Al-Babili, S. Characterization of the rice carotenoid cleavage dioxygenase1 reveals a novel route for geranial biosynthesis. FEBS J. 2009, 276, 736–747. [Google Scholar] [CrossRef]
- Simkin, A.J.; Underwood, B.A.; Auldridge, M.; Loucas, H.M.; Shibuya, K.; Schmelz, E.; Clark, D.G.; Klee, H.J. Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of petunia flowers. Plant Physiol. 2004, 136, 3504–3514. [Google Scholar] [CrossRef]
- Ibdah, M.; Azulay, Y.; Portnoy, V.; Wasserman, B.; Bar, E.; Meir, A.; Burger, Y.; Hirschberg, J.; Schaffer, A.A.; Katzir, N.; et al. Functional characterization of CmCCD1, a carotenoid cleavage dioxygenase from melon. Phytochemistry 2006, 67, 1579–1589. [Google Scholar] [CrossRef]
- Simkin, A.J.; Schwartz, S.H.; Auldridge, M.; Taylor, M.G.; Klee, H.J. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 2004, 40, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Oda-Yamamizo, C.; Sage-Ono, K.; Ohmiya, A.; Ono, M. Alteration of flower colour in Ipomoea nil through CRISPR/Cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 4. Transgenic Res. 2018, 27, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Song, M.H.; Lim, S.H.; Kim, J.K.; Jung, E.S.; John, K.M.M.; You, M.K.; Ahn, S.N.; Lee, C.H.; Ha, S.H. In planta cleavage of carotenoids by Arabidopsis carotenoid cleavage dioxygenase 4 in transgenic rice plants. Plant Biotechnol. Rep. 2016, 10, 291–300. [Google Scholar] [CrossRef]
- Adami, M.; De Franceschi, P.; Brandi, F.; Liverani, A.; Giovannini, D.; Rosati, C.; Dondini, L.; Tartarini, S. Identifying a Carotenoid Cleavage Dioxygenase (ccd4) Gene Controlling Yellow/White Fruit Flesh Color of Peach. Plant Mol. Biol. Rep. 2013, 31, 1166–1175. [Google Scholar] [CrossRef]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Bruno, M.; Beyer, P.; Al-Babili, S. The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of β-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 2015, 572, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.R.; Song, M.H.; Kim, J.K.; Baek, S.A.; You, M.K.; Lim, S.H.; Ha, S.H. RNAi-mediated suppression of three carotenoid-cleavage dioxygenase genes, OsCCD1, 4a, and 4b, increases carotenoid content in rice. J. Exp. Bot. 2018, 69, 5105–5116. [Google Scholar] [CrossRef]
- Baba, S.A.; Jain, D.; Abbas, N.; Ashraf, N. Overexpression of Crocus carotenoid cleavage dioxygenase, CsCCD4b, in Arabidopsis imparts tolerance to dehydration, salt and oxidative stresses by modulating ROS machinery. J. Plant Physiol. 2015, 189, 114–125. [Google Scholar] [CrossRef]
- Butt, H.; Jamil, M.; Wang, J.Y.; Al-Babili, S.; Mahfouz, M. Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Qing, X.Q.; Loewen, M.C. The Biochemical Characterization of Two Carotenoid Cleavage Enzymes from Arabidopsis indicates that a Carotenoid-Derived Compound Inhibits Lateral Branching. J. Biol. Chem. 2004, 279, 46940–46945. [Google Scholar] [CrossRef]
- Hasegawa, S.; Tsutsumi, T.; Fukushima, S.; Okabe, Y.; Saito, J.; Katayama, M.; Shindo, M.; Yamada, Y.; Shimomura, K.; Yoneyama, K.; et al. Low Infection of Phelipanche aegyptiaca in Micro-Tom Mutants Deficient in CAROTENOIDCLEAVAGE DIOXYGENASE 8. Int. J. Mol. Sci. 2018, 19, 2645. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.V.; Leyva-Gonzalez, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Dong, N.V.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. USA 2013, 111, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Jacqueline, T.C.; Jan, A.D.Z. Characterization of the 9-Cis-Epoxycarotenoid Dioxygenase Gene Family and the Regulation of Abscisic Acid Biosynthesis in Avocado. Plant Physiol. 2000, 124, 343–353. [Google Scholar] [CrossRef]
- Luo, P.; Shen, Y.X.; Jin, S.X.; Huang, S.S.; Cheng, X.; Wang, Z.; Li, P.H.; Zhao, J.; Bao, M.Z.; Ning, G.G. Overexpression of Rosa rugosa anthocyanidin reductase enhances tobacco tolerance to abiotic stress through increased ROS scavenging and modulation of ABA signaling. Plant Sci. 2016, 245, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.G.; Zhuo, C.L.; Qian, C.M.; Xiao, T.; Guo, Z.F.; Lu, S.Y. Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 2016, 14, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Stress-Inducible Gene for 9-cis-Epoxycarotenoid Dioxygenase Involved in Abscisic Acid Biosynthesis under Water Stress in Drought-Tolerant Cowpea. Plant Physiol. 2000, 123, 553–562. [Google Scholar] [CrossRef]
- He, R.R.; Zhuang, Y.; Cai, Y.M.; Agüero, C.B.; Liu, S.L.; Wu, J.; Deng, S.H.; Walker, M.A.; Lu, J.; Zhang, Y.L. Overexpression of 9-cis-Epoxycarotenoid Dioxygenase Cisgene in Grapevine Increases Drought Tolerance and Results in Pleiotropic Effects. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef]
- Chen, W.K.; Yu, K.J.; Liu, B.; Lan, Y.B.; Sun, R.Z.; Li, Q.; He, F.; Pan, Q.H.; Duan, C.Q.; Wang, J. Comparison of transcriptional expression patterns of carotenoid metabolism in ‘Cabernet Sauvignon’ grapes from two regions with distinct climate. J. Plant Physiol. 2017, 213, 75–86. [Google Scholar] [CrossRef]
- Yang, Y.X.; Zhou, Q.Q.; Xu, J.Y.; Li, Q.C.; Zhang, S.T. RNA interference of NtNCED3 reduces drought tolerance and impairs plant growth through feedback regulation of isoprenoids in Nicotiana tabacum. Environ. Exp. Bot. 2018, 155, 332–344. [Google Scholar] [CrossRef]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Liu, H.Q.; Guo, Q.W.; Zheng, C.F.; Li, C.S.; Xiang, X.M.; Zhao, D.F.; Liu, J.; Luo, J.; Zhao, D.K.; et al. Genome-wide identification, phylogenetic relationships, and expression analysis of the carotenoid cleavage oxygenase gene family in pepper. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, Z.C.; Ding, A.M.; Kong, Y.Z. Genome-Wide Identification and Expression Profiling Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family in Tobacco (Nicotiana tabacum L.). Genes 2018, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Guo, W.X.; Wu, G.T.; Lu, Y.W.; Peng, J.J.; Zheng, H.Y.; Lin, L.; Chen, J.P. A virus-based miRNA suppression (VbMS) system for miRNA loss-of-function analysis in plants. Biotechnol. J. 2014, 9, 702–708. [Google Scholar] [CrossRef]
- Yin, Z.M.; Murawska, Z.; Xie, F.L.; Pawelkowicz, M.; Michalak, K.; Zhang, B.H.; Lebecka, R. microRNA response in potato virus Y infected tobacco shows strain-specificity depending on host and symptom severity. Virus Res. 2019, 260, 20–32. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; He, Y.H.; Xia, R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Gao, F.L.; Chen, C.J.; Arab, D.A.; Du, Z.G.; He, Y.H.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Species | Gene Name | Gene ID | Gene Length (bp) | ORF Length (bp) | Amino Acid | MW (kDa) | PI | Chromosome | Protein Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| Nicotiana tabacum | NtCCD1a | Ntab0245430 | 8618 | 1641 | 546 | 61.080 | 6.01 | Chr02 | Cytoplasm |
| NtCCD1b | Ntab0755880 | 10472 | 1683 | 560 | 62.932 | 6.51 | Chr06 | Cytoplasm | |
| NtCCD4a | Ntab0608760 | 3600 | 1803 | 600 | 65.858 | 7.16 | Chr09 | Chloroplast | |
| NtCCD4b | Ntab0943160 | 3449 | 1806 | 601 | 65.936 | 7.24 | Chr20 | Chloroplast | |
| NtCCD4c | Ntab0104800 | 2941 | 1803 | 600 | 65.709 | 6.44 | Chr08 | Chloroplast | |
| NtCCD7a | Ntab0292410 | 9135 | 1956 | 650 | 73.341 | 6.43 | Chr13 | Mitochondrion | |
| NtCCD7b | Ntab0552870 | 4928 | 1953 | 651 | 73.260 | 6.43 | Chr06 | Mitochondrion | |
| NtCCD8a | Ntab0551270 | 4366 | 1671 | 556 | 62.350 | 5.98 | Chr18 | Cytoplasm | |
| NtCCD8b | Ntab0369080 | 3707 | 1668 | 555 | 62.194 | 6.18 | Chr06 | Cytoplasm | |
| NtCCDLa | Ntab0667500 | 3102 | 1593 | 530 | 59.969 | 5.85 | Chr06 | Cytoplasm | |
| NtCCDLb | Ntab0429710 | 3221 | 1572 | 523 | 59.214 | 5.56 | Ntab_scaffold_247 | Cytoplasm | |
| NtCCDLc | Ntab0264020 | 3002 | 1470 | 489 | 55.733 | 6.34 | Ntab_scaffold_247 | Cytoplasm | |
| NtNCED2 | Ntab0264020 | 1800 | 1800 | 599 | 66.844 | 7.60 | Ntab_scaffold_176 | Chloroplast | |
| NtNCED3a | Ntab0266320 | 1830 | 1830 | 609 | 67.754 | 7.28 | Chr11 | Cytoplasm | |
| NtNCED3b | Ntab0214800 | 1842 | 1842 | 613 | 68.191 | 7.66 | Chr07 | Cytoplasm | |
| NtNCED5a | Ntab0972510 | 1806 | 1806 | 601 | 67.018 | 6.12 | Chr23 | Chloroplast | |
| NtNCED5b | Ntab0027880 | 1806 | 1806 | 601 | 67.134 | 6.40 | Chr24 | Chloroplast | |
| NtNCED6a | Ntab0557140 | 1795 | 1746 | 581 | 64.607 | 8.18 | Chr16 | Chloroplast | |
| NtNCED6b | Ntab0434870 | 8137 | 1608 | 535 | 60.419 | 4.60 | Chr15 | Cytoplasm | |
| Nicotiana sylvestris | NsyCCD1a | Nsyl0144600 | 13565 | 1707 | 568 | 63.685 | 6.49 | Nsyl_scaffold_1522 | Cytoplasm |
| NsyCCD1b | Nsyl0177010 | 8624 | 1647 | 548 | 61.316 | 6.33 | Nsyl_superscaffold_202 | Cytoplasm | |
| NsyCCD4 | Nsyl0462250 | 2426 | 1803 | 600 | 65.691 | 6.62 | Nsyl_superscaffold_41 | Chloroplast | |
| NsyCCD7 | Nsyl0304670 | 9105 | 1953 | 650 | 73.341 | 6.43 | Nsyl_superscaffold_23 | Mitochondrion | |
| NsyCCD8 | Nsyl0145940 | 4366 | 1671 | 556 | 62.350 | 5.98 | Nsyl_superscaffold_190 | Cytoplasm | |
| NsyCCDL | Nsyl0401980 | 3221 | 1572 | 523 | 59.214 | 5.56 | Nsyl_superscaffold_155 | Cytoplasm | |
| NsyNCED2 | Nsyl0071360 | 1800 | 1800 | 599 | 66.844 | 7.60 | Nsyl_superscaffold_251 | Chloroplast | |
| NsyNCED3 | Nsyl0483670 | 1830 | 1830 | 609 | 67.754 | 7.28 | Nsyl_superscaffold_179 | Cytoplasm | |
| NsyNCED5 | Nsyl0074010 | 1806 | 1806 | 601 | 67.020 | 6.12 | Nsyl_scaffold_1118 | Chloroplast | |
| NsyNCED6 | Nsyl0472000 | 1518 | 1518 | 505 | 56.325 | 5.83 | Nsyl_superscaffold_26 | Cytoplasm | |
| Nicotiana tomentosiformis | NtoCCD1a | Ntom0051870 | 8169 | 1647 | 548 | 61.657 | 6.27 | Ntom_scaffold_148 | Cytoplasm |
| NtoCCD1b | Ntom0051880 | 10899 | 1641 | 546 | 61.138 | 6.06 | Ntom_scaffold_14 | Cytoplasm | |
| NtoCCD4a | Ntom0284420 | 3600 | 1803 | 600 | 65.858 | 7.16 | Ntom_scaffold_554 | Chloroplast | |
| NtoCCD4b | Ntom0290610 | 3458 | 1806 | 601 | 65.940 | 6.58 | Ntom_superscaffold_38 | Chloroplast | |
| NtoCCD7 | Ntom0214430 | 5121 | 1953 | 650 | 73.176 | 6.43 | Ntom_scaffold_374 | Mitochondrion | |
| NtoCCD8 | Ntom0326520 | 3707 | 1668 | 555 | 62.183 | 6.30 | Ntom_scaffold_709 | Cytoplasm | |
| NtoCCDLa | Ntom0006270 | 3289 | 1668 | 555 | 62.702 | 6.34 | Ntom_scaffold_103 | Chloroplast | |
| NtoCCDLb | Ntom0326460 | 3086 | 1563 | 523 | 59.238 | 6.23 | Ntom_scaffold_709 | Cytoplasm | |
| NtoNCED3 | Ntom0303590 | 1842 | 1842 | 613 | 68.177 | 7.66 | Ntom_scaffold_600 | Cytoplasm | |
| NtoNCED5 | Ntom0232040 | 1806 | 1806 | 601 | 67.094 | 6.37 | Ntom_superscaffold_17 | Chloroplast | |
| NtoNCED6 | Ntom0304970 | 1752 | 1752 | 583 | 64.887 | 8.35 | Ntom_superscaffold_63 | Chloroplast |
| miRNA_Acc. | Target_Acc. | Expectation | UPE | miRNA Length | Target Start | Target End | Inhibition | Multiplicity |
|---|---|---|---|---|---|---|---|---|
| nta-miR319a | NtCCDLc | 3 | 21.947 | 21 | 1266 | 1286 | Cleavage | 1 |
| nta-miR319b | NtCCDLc | 3 | 22.68 | 21 | 1265 | 1285 | Cleavage | 1 |
| nta-miR159 | NtCCDLc | 3 | 22.68 | 21 | 1265 | 1285 | Cleavage | 1 |
| nta-miR6150 | NtNCED3a | 3.5 | 10 | 22 | 1433 | 1454 | Cleavage | 1 |
| nta-miR6150 | NtNCED3b | 3.5 | 11.413 | 22 | 1445 | 1466 | Cleavage | 1 |
| nta-miR6150 | NtNCED6a | 4 | 21.187 | 22 | 1112 | 1134 | Translation | 1 |
| nta-miR6148b | NtCCD1a | 4 | 14.183 | 21 | 645 | 665 | Translation | 1 |
| nta-miR6164a | NtCCD1a | 4 | 18.268 | 21 | 356 | 376 | Translation | 1 |
| nta-miR6164b | NtCCD1a | 4 | 18.268 | 21 | 356 | 376 | Translation | 1 |
| nta-miR159 | NtCCD4a | 4 | 8.152 | 21 | 324 | 344 | Translation | 1 |
| Model | Np | Ln L | Estimates of Parameters | RT Pairs | LRT p-Value | Positive Sites |
|---|---|---|---|---|---|---|
| M0: one ratio | 38 | −13,579.23 | ω0 = 0.09246 | |||
| M3: discrete | 42 | −13,413.93 | p0 = 0.14105, p1 = 0.62465, p2 = 0.23430, | M0 vs. M3 | 0.00 | None |
| ω0 = 0.01164, ω1 = 0.06928, ω2 = 0.25309 | ||||||
| M2a: selection | 41 | −13,554.18 | p0 = 0.935245, p1 = 0.03719, p2 = 0.02757, | |||
| ω0 = 0.08829, ω1 = 1.00000, ω2 = 1.00000 | ||||||
| M1a: neutral | 39 | −13,554.18 | p0 = 0.93524, p1 = 0.06476, | M1a vs. M2a | 1.00 | None |
| ω0 = 0.08829, ω1 = 1.00000 | ||||||
| M8: beta and ω > 1 | 41 | −13,430.72 | p0 = 0.99883, p = 1.12281, q = 9.15782, | M7 vs. M8 | 0.9839 | None |
| p1 = 0.00117, ω = 1.00000 | ||||||
| M7: beta | 39 | −13,430.73 | p = 1.11747, q = 9.06492 | |||
| M8a: beta and ω = 1 | 40 | −13,419.70 | p0 = 0.99999, p = 1.35368, q = 11.54435, | M8a vs. M8 | 0.000002674 | None |
| p1 = 0.0000, ω = 1.00000 | ||||||
| Model | Np | Ln L | Estimates of Parameters | LRT p-Value | Positive Sites (BEB) |
|---|---|---|---|---|---|
| Model 0 | 38 | −13579.23 | ω = 0.09246 | ||
| Two ratio branch a | 39 | −13575.32 | ω0 = 0.10005, ω1 = 0.04060 | 0.005 | None |
| Two ratio branch b | 39 | −13564.78 | ω0 = 0.10374, ω1 = 0.00918 | 0.000 | None |
| Two ratio branch c | 39 | −13574.47 | ω0 = 0.09358, ω1 = 0.00164 | 0.002 | None |
| Two ratio branch d | 39 | −13574.37 | ω0 = 0.09763, ω1 = 0.02200 | 0.002 | None |
| Two ratio branch e | 39 | −13574.07 | ω0 = 0.09443, ω1 = 0.00421 | 0.001 | None |
| Two ratio branch f | 39 | −13578.74 | ω0 = 0.09401, ω1 = 0.04312 | 0.321 | None |
| Two ratio branch g | 39 | −13577.44 | ω0 = 0.09348, ω1 = 0.00024 | 0.059 | None |
| Two ratio branch h | 39 | −13574.78 | ω0 = 0.09439, ω1 = 0.00432 | 0.003 | None |
| Two ratio branch i | 39 | −13572.92 | ω0 = 0.09489, ω1 = 0.00216 | 0.00 | None |
| Two ratio branch j | 39 | −13579.19 | ω0 = 0.09241, ω1 = 0.12516 | 0.772 | None |
| Two ratio branch k | 39 | −13578.89 | ω0 = 0.09278, ω1 = 0.00154 | 0.409 | None |
| Two ratio branch l | 39 | −13579.14 | ω0 = 0.09277, ω1 = 0.05689 | 0.662 | None |
| Two ratio branch m | 39 | −13578.69 | ω0 = 0.09311, ω1 = 0.00201 | 0.297 | None |
| Two ratio branch n | 39 | −13576.50 | ω0 = 0.09367, ω1 = 0.00833 | 0.019 | None |
| Model | Np | Ln L | Estimates of Parameters | LRT p-Value | Positive Sites (BEB) |
|---|---|---|---|---|---|
| Model A (Branch g) | 41 | −13551.77 | p0 = 0.90022, p1 = 0.06108, p2a = 0.03624, p2b = 0.00246, ω0 = 0.08803, ω1 = 1.00000, ω2a = 999.00000, ω2b = 999.00000 | 0.03 | None |
| Model A Null (Branch g) | 40 | −13554.00 | p0 = 0.86534, p1 = 0.05982, p2a = 0.07000, p2b =0.00484, ω0 = 0.08808, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | Not Allowed | |
| Model A (Branch i) | 41 | −13545.47 | p0 = 0.81697, p1 = 0.05118, p2a = 0.12408, p2b = 0.00777, ω0 = 0.08849, ω1 = 1.00000, ω2a = 11.81766, ω2b = 11.81766 | 0.01 | 131K *, 339I **, 343A *, 349G *, 406F *, 413Y *, 533F * |
| Model A Null (Branch i) | 40 | −13548.71 | p0 = 0.79861, p1 = 0.05219, p2a = 0.14005, p2b = 0.00915, ω0 = 0.0867, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | Not Allowed | |
| Model A (Branch k) | 41 | −13545.83 | p0 = 0.79074, p1 = 0.04952, p2a = 0.15032, p2b = 0.00941, ω0 = 0.08852, ω1 = 1.00000, ω2a = 999.00000, ω2b = 999.00000 | 0.0009 | None |
| Model A Null (Branch g) | 40 | −13551.29 | p0 = 0.77180, p1 = 0.05151, p2a = 0.16563, p2b = 0.01105, ω0 = 0.08705, ω1 = 1.00000, ω2a = 1.00000, ω2b = 1.00000 | Not Allowed |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Li, Q.; Li, P.; Zhang, S.; Liu, C.; Jin, J.; Cao, P.; Yang, Y. Carotenoid Cleavage Dioxygenases: Identification, Expression, and Evolutionary Analysis of This Gene Family in Tobacco. Int. J. Mol. Sci. 2019, 20, 5796. https://doi.org/10.3390/ijms20225796
Zhou Q, Li Q, Li P, Zhang S, Liu C, Jin J, Cao P, Yang Y. Carotenoid Cleavage Dioxygenases: Identification, Expression, and Evolutionary Analysis of This Gene Family in Tobacco. International Journal of Molecular Sciences. 2019; 20(22):5796. https://doi.org/10.3390/ijms20225796
Chicago/Turabian StyleZhou, Qianqian, Qingchang Li, Peng Li, Songtao Zhang, Che Liu, Jingjing Jin, Peijian Cao, and Yongxia Yang. 2019. "Carotenoid Cleavage Dioxygenases: Identification, Expression, and Evolutionary Analysis of This Gene Family in Tobacco" International Journal of Molecular Sciences 20, no. 22: 5796. https://doi.org/10.3390/ijms20225796
APA StyleZhou, Q., Li, Q., Li, P., Zhang, S., Liu, C., Jin, J., Cao, P., & Yang, Y. (2019). Carotenoid Cleavage Dioxygenases: Identification, Expression, and Evolutionary Analysis of This Gene Family in Tobacco. International Journal of Molecular Sciences, 20(22), 5796. https://doi.org/10.3390/ijms20225796