HUNK Phosphorylates Rubicon to Support Autophagy
Abstract
1. Introduction
2. Results
2.1. HUNK Kinase Activity is Sufficient for Autophagy
2.2. HUNK Binds the Beclin-1 Protein Complex
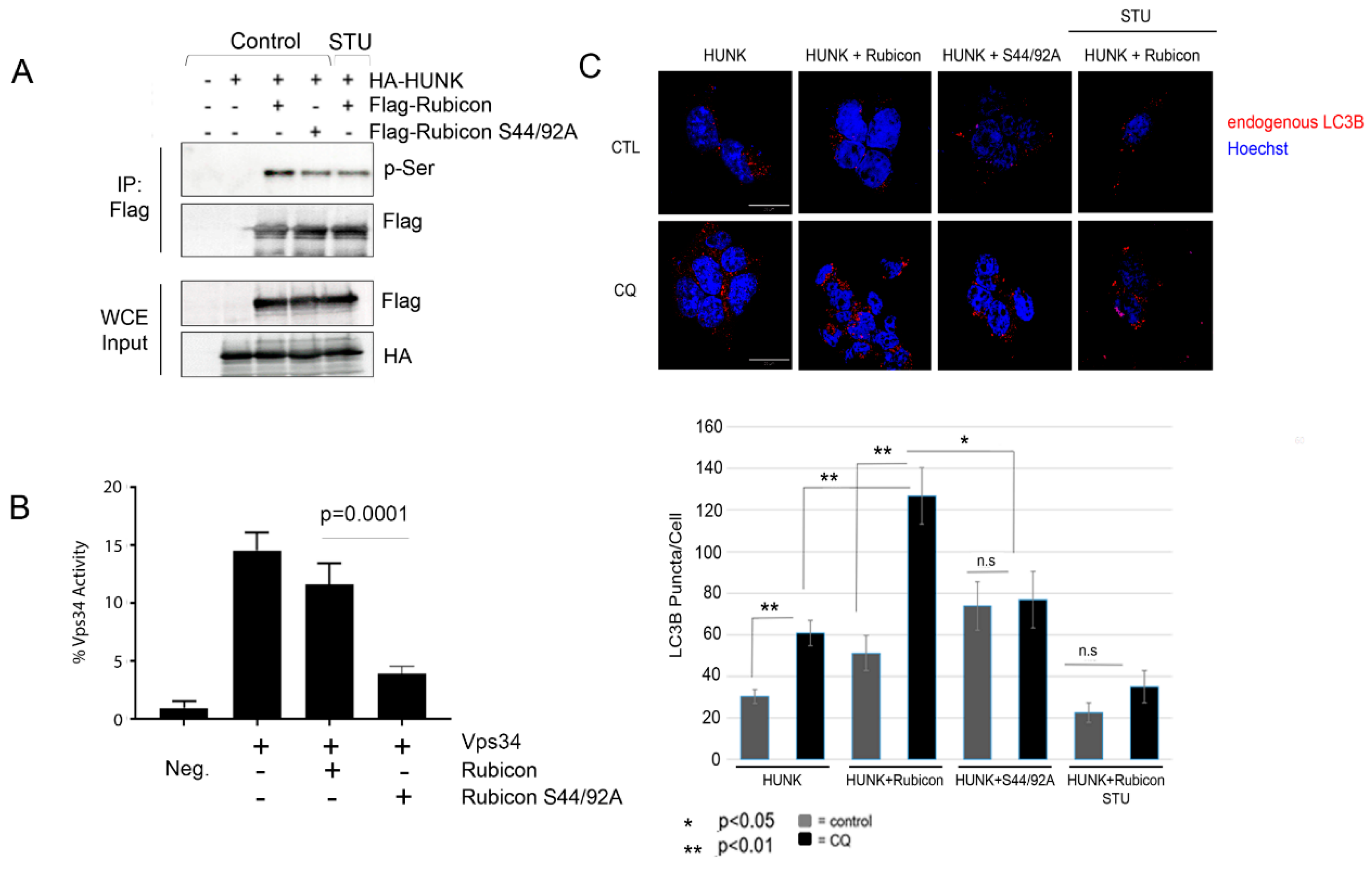
2.3. HUNK Phosphorylates Rubicon within the N-terminus of the Protein
2.4. HUNK Phosphorylation of Rubicon Promotes Autophagosome Formation
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Immunoblotting
4.3. Immunofluorescence
4.4. Immunoprecipitation Assay
4.5. HUNK Kinase Assay
4.6. Vps34 Kinase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Beclin-1 | Coiled-coil myosin-like BCL2-interacting protein |
| Vps34 | Vacuolar protein sorting, class III PI 3-kinase |
| Atg14L | Autophagy related 14 like |
| UVRAG | UV radiation resistance-associated gene protein |
| Rubicon | RUN domain Beclin-1-interacting and cysteine-rich domain-containing protein |
| AMPK | AMP-activated protein kinase |
| mTOR | Mammalian target of rapamycin |
| Akt | Protein kinase B |
| ULK1 | Unc-51 like autophagy activating kinase 1 |
| EGFR | Epidermal growth factor receptor |
| HUNK | Hormonally upregulated neu-associated kinase |
| LC3B | Microtubule-associated proteins 1A/1B light chain 3B |
| WT | Wildtype |
| RFP | Red fluorescent protein |
| GFP | Green fluorescent protein |
| CQ | Chloroquine |
| RUN | RPIP8, unc-14, and NESCA domain |
| STU | Staurosporine |
| GST | Glutathione S-transferase |
| DMSO | Dimethyl sulfoxide |
| Rab7 | Ras-related protein 7 |
| GDP | Guanosine diphosphate |
| GTP | Guanosine triphosphate |
| HER2 | Human epidermal growth factor receptor 2 |
References
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdalla, F.C.; Abeliovich, H.; Abraham, R.T.; Acevedo-Arozena, A.; Adeli, K.; Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-Ghiso, J.A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012, 8, 445–544. [Google Scholar] [PubMed]
- Klionsky, D.J.; Abeliovich, H.; Agostinis, P.; Agrawal, D.K.; Aliev, G.; Askew, D.S.; Baba, M.; Baehrecke, E.H.; Bahr, B.A.; Ballabio, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008, 4, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Cheng, Y.; Liu, B. Beclin-1: Autophagic regulator and therapeutic target in cancer. Int. J. Biochem. Cell Biol. 2013, 45, 921–924. [Google Scholar] [CrossRef]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef]
- Matsunaga, K.; Noda, T.; Yoshimori, T. Binding Rubicon to cross the Rubicon. Autophagy 2009, 5, 876–877. [Google Scholar] [CrossRef]
- McKnight, N.C.; Zhenyu, Y. Beclin 1, an Essential Component and Master Regulator of PI3K-III in Health and Disease. Curr. Pathobiol. Rep. 2013, 1, 231–238. [Google Scholar] [CrossRef]
- Menon, M.B.; Dhamija, S. Beclin 1 Phosphorylation—At the Center of Autophagy Regulation. Front. Cell Dev. Biol. 2018, 6, 137. [Google Scholar] [CrossRef]
- Sridharan, S.; Jain, K.; Basu, A. Regulation of autophagy by kinases. Cancers 2011, 3, 2630–2654. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.C.; Fang, C.; Russell, R.C.; Kim, J.H.; Fan, W.; Liu, R.; Zhong, Q.; Guan, K.L. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013, 152, 290–303. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, W.; Sun, X.; Xu, D.; Wang, C.; Zhang, Q.; Wang, H.; Luo, W.; Chen, Y.; Chen, H.; et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy 2016, 12, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zou, Z.; Becker, N.; Anderson, M.; Sumpter, R.; Xiao, G.; Kinch, L.; Koduru, P.; Christudass, C.S.; Veltri, R.W.; et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013, 154, 1269–1284. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Phelps-Polirer, K.; Abt, M.A.; Smith, D.; Yeh, E.S. Co-Targeting of JNK and HUNK in Resistant HER2-Positive Breast Cancer. PLoS ONE 2016, 11, e0153025. [Google Scholar] [CrossRef]
- Yeh, E.S.; Abt, M.A.; Hill, E.G. Regulation of cell survival by HUNK mediates breast cancer resistance to HER2 inhibitors. Breast Cancer Res. Treat. 2015, 149, 91–98. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, J.; Fan, W.; Wong, K.N.; Ding, X.; Chen, S.; Zhong, Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J. Biol. Chem. 2011, 286, 185–191. [Google Scholar] [CrossRef]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Zambrano, J.; Williams, C.J.; Williams, C.B.; Hedgepeth, L.; Burger, P.; Dilday, T.; Eblen, S.T.; Armeson, K.; Hill, E.G.; Yeh, E.S. Staurosporine, an inhibitor of hormonally up-regulated neu-associated kinase. Oncotarget 2018, 9, 35962–35973. [Google Scholar] [CrossRef]
- Williams, C.B.; Phelps-Polirer, K.; Dingle, I.P.; Williams, C.J.; Rhett, J.M.; Eblen, S.T.; Armeson, K.; Hill, E.G.; Yeh, E.S. HUNK Phosphorylates EGFR to Regulate Breast Cancer Metastasis. Oncogene 2019. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Westphal, W.; Wong, K.N.; Tan, I.; Zhong, Q. Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. USA 2010, 107, 19338–19343. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Lee, J.S.; Inn, K.S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zambrano, J.N.; Eblen, S.T.; Abt, M.; Rhett, J.M.; Muise-Helmericks, R.; Yeh, E.S. HUNK Phosphorylates Rubicon to Support Autophagy. Int. J. Mol. Sci. 2019, 20, 5813. https://doi.org/10.3390/ijms20225813
Zambrano JN, Eblen ST, Abt M, Rhett JM, Muise-Helmericks R, Yeh ES. HUNK Phosphorylates Rubicon to Support Autophagy. International Journal of Molecular Sciences. 2019; 20(22):5813. https://doi.org/10.3390/ijms20225813
Chicago/Turabian StyleZambrano, Joelle N., Scott T. Eblen, Melissa Abt, J. Matthew Rhett, Robin Muise-Helmericks, and Elizabeth S. Yeh. 2019. "HUNK Phosphorylates Rubicon to Support Autophagy" International Journal of Molecular Sciences 20, no. 22: 5813. https://doi.org/10.3390/ijms20225813
APA StyleZambrano, J. N., Eblen, S. T., Abt, M., Rhett, J. M., Muise-Helmericks, R., & Yeh, E. S. (2019). HUNK Phosphorylates Rubicon to Support Autophagy. International Journal of Molecular Sciences, 20(22), 5813. https://doi.org/10.3390/ijms20225813





