Identification of Flower-Specific Promoters through Comparative Transcriptome Analysis in Brassica napus
Abstract
:1. Introduction
2. Results
2.1. Transcriptome Analysis and Identification of Tissue-Specific Genes
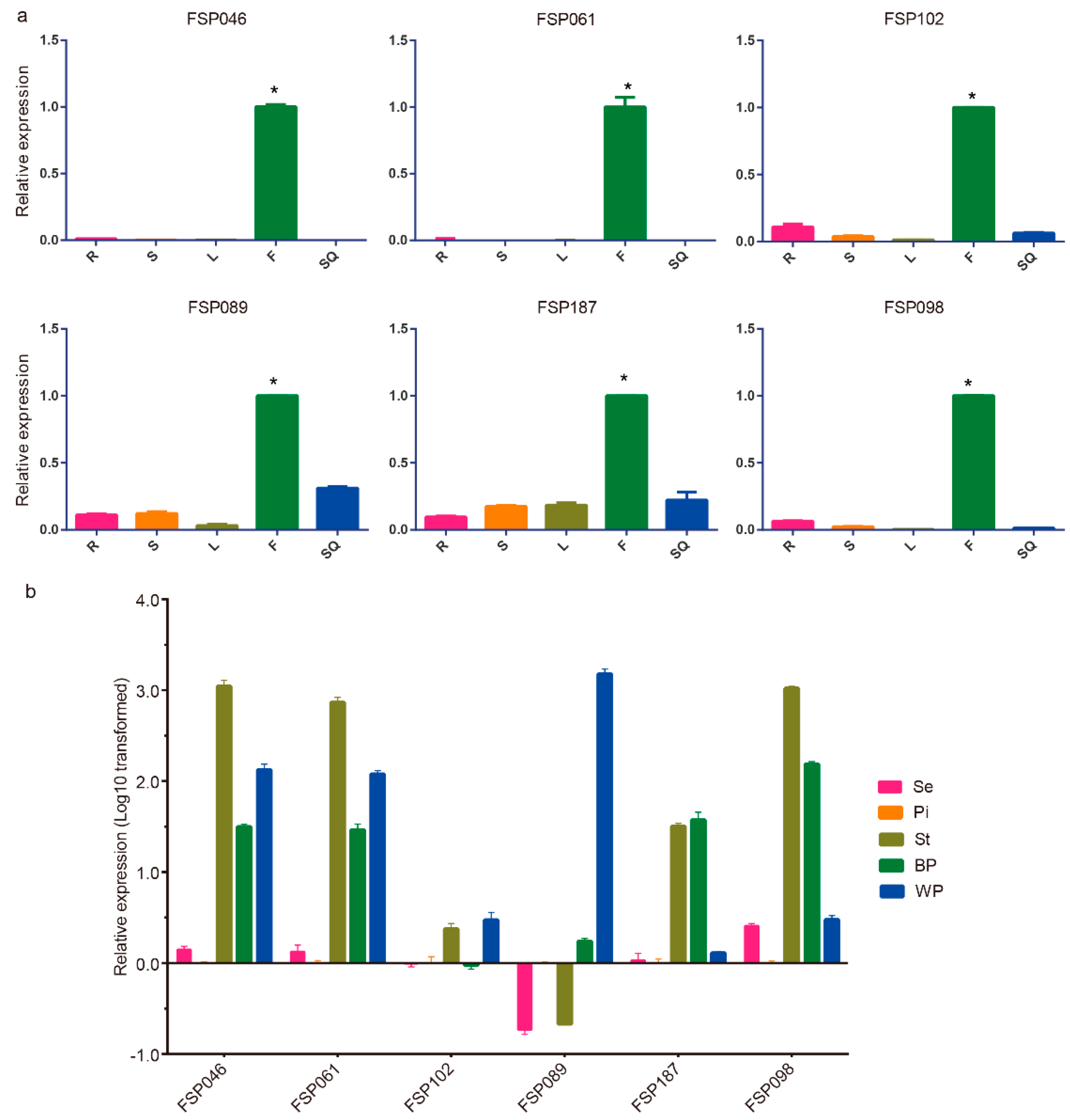
2.2. qRT-PCR Analysis of Flower-Specific Candidate Genes
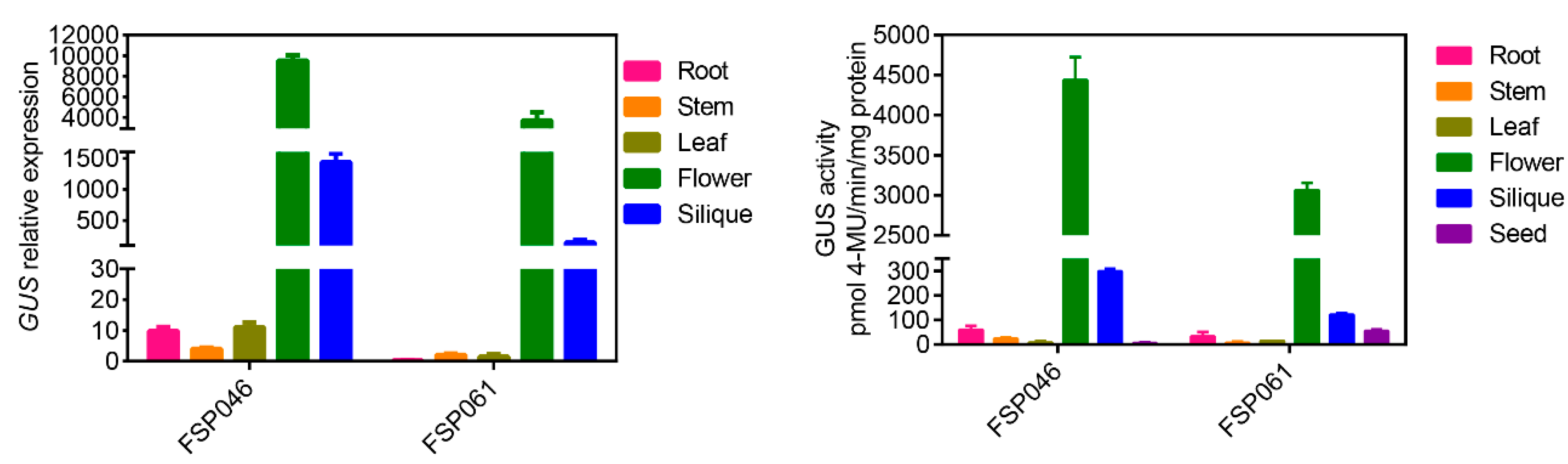
2.3. FSP046 and FSP061 Promoters are Flower-Specific in Transgenic A. thaliana
2.4. FSP046 and FSP061 promoter can Drive GUS Expressing in Flowers of B. napus
3. Discussion
4. Materials and Methods
4.1. Plant Material Collection
4.2. RNA Preparation and RNA-seq Analysis
4.3. Functional Annotation
4.4. Screening of Tissue-Specific Candidate Genes
4.5. Quantitative Real-Time PCR Analysis
4.6. Cloning and Characterization of Flower-Specific Promoters
4.7. Binary Vector Construction and Plant Transformation
4.8. Histochemical GUS Assays and Evaluation of GUS Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| SSR | Sclerotinia stem rot |
| BP | Blossomy petal |
| WP | Wilting petal |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| GO | Gene Ontology |
| Qrt-PCR | Quantitative real-time PCR |
| 4-MU | 4-methylumbelliferone |
References
- Chalhoub, B.; Denoeud, F.; Liu, S.Y.; Parkin, I.A.P.; Tang, H.B.; Wang, X.Y.; Chiquet, J.; Belcram, H.; Tong, C.B.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Wang, J.; Lydiate, D.J.; Parkin, I.A.P.; Falentin, C.; Delourme, R.; Carion, P.W.C.; King, G.J. Integration of linkage maps for the Amphidiploid Brassica napus and comparative mapping with Arabidopsis and Brassica rapa. BMC Genom. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant. Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Bolton, M.D.; Thomma, B.P.H.J.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.M.; Savocchia, S.; Lindbeck, K.D.; Ash, G.J. Biology and biocontrol of Sclerotinia sclerotiorum (Lib.) de Bary in oilseed Brassicas. Australa. Plant Path. 2016, 45, 1–14. [Google Scholar] [CrossRef]
- Liang, X.; Rollins, J.A. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140. [Google Scholar] [CrossRef]
- Aggarwal, R.A.K.; Kumar, A.; Thakur, H.L. Effect of Sclerotinia rot on oil quality in low erucic acid cultivars of rapeseed. Cruciferae Newsl. 1997, 19, 103–104. [Google Scholar]
- Turkington, T.K.; Raa, M. Use of petal infestation to forecast Sclerotinia stem rot of canola: The influence of inoculum variation over the flowering period and canopy density. Phytopathology 1993, 83, 561–565. [Google Scholar] [CrossRef]
- Inglis, G.D.; Boland, G.J. The microflora of bean and rapeseed petals and the influence of the microflora of bean petals on white mold. Can. J. Plant Pathol. 1990, 2, 129–134. [Google Scholar] [CrossRef]
- Saharan, G.S.; Mehta, N. Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management, 1st ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 71–75. [Google Scholar]
- Van der Biezen, E.A. Quest for antimicrobial genes to engineer disease-resistant crops. Trends Plant Sci. 2001, 6, 89–91. [Google Scholar] [CrossRef]
- Shikata, M.; Narumi, T.; Yamaguchi, H.; Sasaki, K.; Aida, R.; Oshima, Y.; Takiguchi, Y.; Ohme-Takagi, M.; Mitsuda, N.; Ohtsubo, N. Efficient production of novel floral traits in torenia by collective transformation with chimeric repressors of Arabidopsis transcription factors. Plant Biotechnol.-Nar 2011, 28, 189–199. [Google Scholar] [CrossRef]
- Pino, M.T.; Skinner, J.S.; Park, E.J.; Jeknić, Z.; Hayes, P.M.; Thomashow, M.F.; Chen, T.H. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnol. J. 2010, 5, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, O.; Masahito, S.; Tomotsugu, K.; Norihiro, O.; Nobutaka, M.; Masaru, O.T. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 2013, 25, 1609–1624. [Google Scholar]
- Hsieh, T.H.; Lee, J.T.; Charng, Y.Y.; Chan, M.T. Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol. 2002, 130, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Zavallo, D.; Bilbao, M.L.; Hopp, H.E.; Heinz, R. Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L.). Plant Cell Rep. 2010, 29, 239–248. [Google Scholar] [CrossRef]
- Hou, J.; Jiang, P.; Qi, S.; Ke, Z.; He, Q.; Xu, C.; Ding, Z.; Zhang, K.; Li, K. Isolation and Functional Validation of Salinity and Osmotic Stress Inducible Promoter from the Maize Type-II H+-Pyrophosphatase Gene by Deletion Analysis in Transgenic Tobacco Plants. Plos ONE 2016, 11, e0154041. [Google Scholar] [CrossRef]
- Zhang, H.; Jing, R.; Mao, X. Functional Characterization of TaSnRK2.8 Promoter in Response to Abiotic Stresses by Deletion Analysis in Transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 1198. [Google Scholar] [CrossRef]
- Liu, Y.L.; Lou, Q.; Xu, W.R.; Xin, Y.; Bassett, C.; Wang, Y.J. Characterization of a chalcone synthase (CHS) flower-specific promoter from Lilium orential ‘Sorbonne’. Plant Cell Rep. 2011, 30, 2187–2194. [Google Scholar] [CrossRef]
- Sasaki, K.; Yamaguchi, H.; Narumi, T.; Shikata, M.; Oshima, Y.; Nakata, M.; Mitsuda, N.; Ohme-Takagi, M.; Ohtsubo, N. Utilization of a floral organ-expressing AP1 promoter for generation of new floral traits in Torenia fournieri Lind. Plant. Biotech.-Nar. 2011, 28, 181–188. [Google Scholar] [CrossRef]
- An, N.U.; Hu, Y.; Li, P.; Yuchi, Z.; Chen, Y.; Zhang, Y. Cloning and expression of a non-ribosomal peptide synthetase to generate blue rose. ACS Synth Biol. 2019, 8, 1698–1704. [Google Scholar] [CrossRef]
- Ma, N.; Chen, W.; Fan, T.; Tian, Y.; Zhang, S.; Zeng, D.; Li, Y. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol. 2015, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar] [PubMed]
- Hill, J.P.; Lord, E.M. Floral development in Arabidopsis thaliana: Comparison of the wildtype and the homeotic pistillata mutant. Can. J. Bot. 1989, 67, 2922–2936. [Google Scholar] [CrossRef]
- Jack, T.; Brockman, L.L.; Meyerowitz, E.M. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 1992, 68, 683–697. [Google Scholar] [CrossRef]
- Hill, T.A.; Day, C.D.; Zondlo, S.C.; Thackeray, A.G.; Irish, V.F. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 1998, 125, 1711–1721. [Google Scholar]
- Honma, T.; Goto, K. The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 2000, 127, 2021–2030. [Google Scholar]
- Roh, K.H.; Choi, S.B.; Kang, H.C.; Kim, J.B.; Kim, H.U.; Lee, K.R.; Sun, H.K. Isolation and functional characterization of a PISTILLATA-1 gene promoter from Brassica napus. J. Korean Soc. Appl. Biol. 2014, 57, 759–768. [Google Scholar] [CrossRef]
- Wan, L.; Xia, X.; Hong, D.; Yang, G. Molecular analysis and expression of a floral organ-specific polygalacturonase gene isolated from rapeseed (Brassica napus L.). Mol. Biol. Rep. 2010, 37, 3851–3862. [Google Scholar] [CrossRef]
- Chiou, C.Y.; Wu, K.Q.; Yeh, K.W. Characterization and promoter activity of chromoplast specific carotenoid associated gene (CHRC) from Oncidium Gower Ramsey. Biotechnol. Lett. 2008, 30, 1861–1866. [Google Scholar] [CrossRef]
- Diatchenko, L.; Lau, Y.F.; Campbell, A.P.; Chenchik, A.; Moqadam, F.; Huang, B.; Lukyanov, S.; Lukyanov, K.; Gurskaya, N.; Sverdlov, E.D. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 1996, 93, 6025–6030. [Google Scholar] [CrossRef]
- Kononen, J.; Bubendorf, L.; Kallioniemi, A.; Barlund, M.; Schraml, P.; Leighton, S.; Torhorst, J.; Mihatsch, M.J.; Sauter, G.; Kallioniemi, O.P. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 1998, 4, 844–847. [Google Scholar] [CrossRef] [PubMed]
- ‘t Hoen, P.A.C.; Ariyurek, Y.; Thygesen, H.H.; Vreugdenhil, E.; Vossen, R.H.A.M.; de Menezes, R.X.; Boer, J.M.; van Ommen, G.J.B.; den Dunnen, J.T. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008, 36, e141. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Kim, H.T.; Hwang, I.; Han, K.H. Tissue-type-specific transcriptome analysis identifies developing xylem-specific promoters in poplar. Plant Biotechnol. J. 2012, 10, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Geng, L.; Duan, X.; Liang, C.; Shu, C.; Song, F.; Zhang, J. Mining tissue-specific contigs from peanut (Arachis hypogaea L.) for promoter cloning by deep transcriptome sequencing. Plant Cell Physiol. 2014, 55, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Meer, I.M.V.D.; Spelt, C.E.; Mol, J.N.M.; Stuitje, A.R. Promoter analysis of the chalcone synthase (chs A) gene of Petunia hybrida: A 67 bp promoter region directs flower-specific expression. Plant Mol. Biol. 1990, 15, 95–109. [Google Scholar] [CrossRef]
- Sablowski, R.W.; Moyano, E.; Culianez-Macia, F.A.; Schuch, W.; Martin, C.; Bevan, M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994, 13, 128–137. [Google Scholar] [CrossRef]
- Han, Y.Y.; Ming, F.; Wang, J.W.; Ye, M.M.; Shen, D.L. A novel chalcone synthase gene from Phalaenopsis orchid that alters floral morphology in transgenic tobacco plants. Plant Mol. Bio. Rep. 2005, 23, 193–194. [Google Scholar] [CrossRef]
- Brocard, I.; Charlot, F.; Teoule, E.; Guerche, P. Petal Specific Promoter and Method for Obtaining Plants Having (P). European: Institut National De La Recherche Agronomique. JP 2001517450-A 309-0CT-2001, 2001. [Google Scholar]
- Fu, W.; Chen, D.; Pan, Q.; Li, F.; Zhao, Z.; Ge, X.; Li, Z. Production of red-flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 2018, 16, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Azuma, M.; Morimoto, R.; Hirose, M.; Morita, Y.; Hoshino, A.; Iida, S.; Oshima, Y.; Mitsuda, N.; Ohme-Takagi, M.; Shiratake, K. A petal-specific InMYB1 promoter from Japanese morning glory: A useful tool for molecular breeding of floricultural crops. Plant Biotechnol. J. 2016, 14, 354–363. [Google Scholar] [CrossRef]
- Young, C.S.; Werner, C.P. Infection routes for Sclerotinia sclerotiorum in apetalous and fully petalled winter oilseed rape. Plant Pathol. 2012, 61, 730–738. [Google Scholar] [CrossRef]
- Dong-Hui, F.U.; Jiang, L.Y.; Mason, A.S.; Xiao, M.L.; Zhu, L.R.; Li-Zhi, L.I.; Zhou, Q.H.; Shen, C.J.; Huang, C.H. Research progress and strategies for multifunctional rapeseed: A case study of China. J. Integr. Agric. 2016, 15, 1673–1684. [Google Scholar]
- Doring, T.F.; Skellern, M.; Watts, N.; Cook, S.M. Colour choice behaviour in the pollen beetle Meligethes aeneus (Coleoptera: Nitidulidae). Physiol. Entomol. 2012, 37, 360–378. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.C.; Miao, X.X. Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013, 20, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Lilley, C.J.; Davies, L.J.; Urwin, P.E. RNA interference in plant parasitic nematodes: A summary of the current status. Parasitology 2012, 139, 630–640. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, H.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [Green Version]
- Cai, Q.; He, B.; Kogel, K.H.; Jin, H. Cross-kingdom RNA trafficking and environmental RNAi – nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 2018, 46, 58–64. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. Plos ONE 2012, 7. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.P.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.C.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.P.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.C.; Gao, G. An Arabidopsis Transcriptional Regulatory Map Reveals Distinct Functional and Evolutionary Features of Novel Transcription Factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.M.; Fan, G.Y.; Hu, Q.; Zhou, Y.M.; Guan, M.; Tong, C.B.; Li, J.N.; Du, D.Z.; Qi, C.K.; Jiang, L.C.; et al. The high-quality genome of Brassica napus cultivar ‘ZS11′ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sung, Y.C.; Lin, C.P.; Chen, J.C. Optimization of virus-induced gene silencing in Catharanthus roseus. Plant Pathol. 2015, 63, 1159–1167. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar]
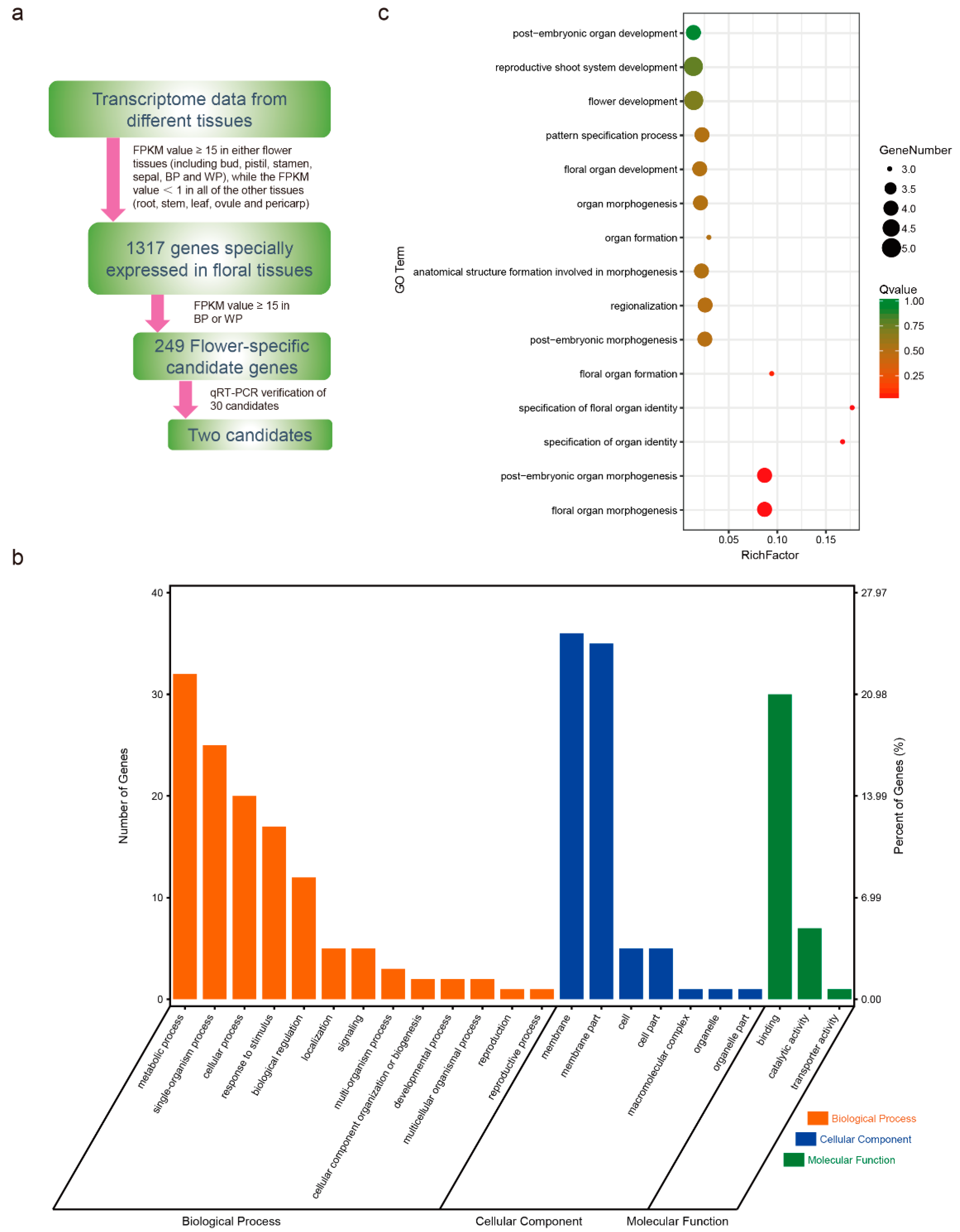



| Root | Leaf | Bud | Silique | Stamen | Pistil | Blossomy Petal * | Wilting Petal ** | Stem | Sepal | Ovule | Pericarp | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clean data | 47,855,967 | 51,669,765 | 50,250,950 | 47,583,789 | 20,822,443 | 23,301,853 | 24,872,044 | 23,779,169 | 18,987,585 | 25,334,038 | 23,375,360 | 25,707,408 |
| All data mapping to genome | 34,126,090 | 35,693,473 | 34,969,636 | 32,285,601 | 18,163,417 | 20,477,668 | 20,852,722 | 17,021,129 | 16,418,565 | 21,868,342 | 20,091,122 | 18,195,703 |
| The percent of all data mapping to genome | 71.31% | 69.08% | 69.59% | 67.85% | 87.23% | 87.88% | 83.84% | 71.58% | 86.47% | 86.32% | 85.95% | 70.78% |
| Unique mapping data | 25,997,362 | 26,362,597 | 25,777,940 | 23,914,692 | 15,536,386 | 17,602,364 | 17,884,295 | 12,917,635 | 14,049,829 | 18,288,123 | 17,027,689 | 13,331,423 |
| The percent of unique mapped data | 54.32% | 51.02% | 51.30% | 50.26% | 74.61% | 75.54% | 71.91% | 54.32% | 73.99% | 72.19% | 72.84% | 51.86% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Dong, C.; Hu, M.; Bai, Z.; Tong, C.; Zuo, R.; Liu, Y.; Cheng, X.; Cheng, M.; Huang, J.; et al. Identification of Flower-Specific Promoters through Comparative Transcriptome Analysis in Brassica napus. Int. J. Mol. Sci. 2019, 20, 5949. https://doi.org/10.3390/ijms20235949
Li Y, Dong C, Hu M, Bai Z, Tong C, Zuo R, Liu Y, Cheng X, Cheng M, Huang J, et al. Identification of Flower-Specific Promoters through Comparative Transcriptome Analysis in Brassica napus. International Journal of Molecular Sciences. 2019; 20(23):5949. https://doi.org/10.3390/ijms20235949
Chicago/Turabian StyleLi, Yan, Caihua Dong, Ming Hu, Zetao Bai, Chaobo Tong, Rong Zuo, Yueying Liu, Xiaohui Cheng, Mingxing Cheng, Junyan Huang, and et al. 2019. "Identification of Flower-Specific Promoters through Comparative Transcriptome Analysis in Brassica napus" International Journal of Molecular Sciences 20, no. 23: 5949. https://doi.org/10.3390/ijms20235949
APA StyleLi, Y., Dong, C., Hu, M., Bai, Z., Tong, C., Zuo, R., Liu, Y., Cheng, X., Cheng, M., Huang, J., & Liu, S. (2019). Identification of Flower-Specific Promoters through Comparative Transcriptome Analysis in Brassica napus. International Journal of Molecular Sciences, 20(23), 5949. https://doi.org/10.3390/ijms20235949





