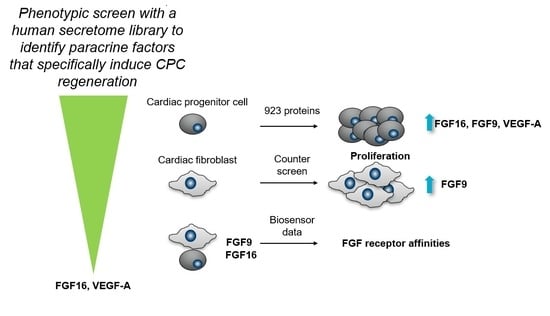
Phenotypic Screen with the Human Secretome Identifies FGF16 as Inducing Proliferation of iPSC-Derived Cardiac Progenitor Cells
Abstract
:1. Introduction
2. Results
2.1. FGF9, FGF16, and 10 Additional Secreted Proteins Identified to Stimulate CPC Proliferation in a Phenotypic Screen
2.2. FGF16, Noggin, NPTX1, and VEGF-A Specifically Induce Proliferation of Human Cardiac Progenitor Cells but not Human Cardiac Fibroblasts
2.3. Biosensor Analysis Suggests That FGF9 and FGF16 Bind to Different Receptors on hCPCs and hCFs
2.4. FGF16 Induces Proliferation of Naïve CPCs Isolated from Mouse Hearts and Human iPS-Derived Cardiomyocytes
3. Discussion
4. Materials and Methods
4.1. Selection of Protein Genes for Inclusion in Secretome Library
4.2. Protein Production and Quality Control
4.3. Sample Management of Proteins
4.4. Human iPSC-Derived CPC Culture and Proliferation Screen
4.5. Human Native Cardiac Fibroblasts Culture and Proliferation Assay
4.6. Isolation of Mouse Sca1-Positive Progenitor Cells from Naïve Hearts and Proliferation Assay
4.7. Human iPSC-Derived Cardiomyocyte Culture and Proliferation Screen
4.8. QCM Experiments
4.9. qPCR Experiments on hCFs and hCPCs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MI | Myocardial infarction |
| CPC | Cardiac progenitor cells |
| iPS-CPC | Induced pluripotent cardiac progenitor cells |
| FGF | Fibroblast growth factor |
| CF | Cardiac fibroblast |
| QCM | Quartz crystal microbalance |
| COP-1 | Cell-optimized polystyrene |
| CR | Concentration response |
| EC | Effective concentration |
References
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; Howard, V.J.; et al. Heart disease and stroke statistics--2015 update: A report from the American Heart Association. Circulation 2015, 131, e29–e322. [Google Scholar] [CrossRef]
- Oberpriller, J.O.; Oberpriller, J.C. Response of the adult newt ventricle to injury. J. Exp. Zool. 1974, 187, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart regeneration in zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef] [PubMed]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H.A. Transient regenerative potential of the neonatal mouse heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef]
- Hashimoto, H.; Olson, E.N.; Bassel-Duby, R. Therapeutic approaches for cardiac regeneration and repair. Nat. Rev. Cardiol. 2018, 15, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, L.; Clarke, J.C.; Yen, C.; Gregoire, F.; Albery, T.; Billger, M.; Egnell, A.C.; Gan, L.M.; Jennbacken, K.; Johansson, E.; et al. Biocompatible, Purified VEGF-A mRNA Improves Cardiac Function after Intracardiac Injection 1 Week Post-myocardial Infarction in Swine. Mol. Ther. Methods Clin. Dev. 2018, 9, 330–346. [Google Scholar] [CrossRef] [PubMed]
- Chien, K.R.; Frisen, J.; Fritsche-Danielson, R.; Melton, D.A.; Murry, C.E.; Weissman, I.L. Regenerating the field of cardiovascular cell therapy. Nat. Biotechnol. 2019, 37, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.M.A.; Ang, Y.S.; Radzinsky, E.; Zhou, P.; Huang, Y.; Elfenbein, A.; Foley, A.; Magnitsky, S.; Srivastava, D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 2018, 173, 104–116. [Google Scholar] [CrossRef]
- He, L.; Han, M.; Zhang, Z.; Li, Y.; Huang, X.; Liu, X.; Pu, W.; Zhao, H.; Wang, Q.D.; Nie, Y.; et al. Reassessment of c-Kit(+) Cells for Cardiomyocyte Contribution in Adult Heart. Circulation 2019, 140, 164–166. [Google Scholar] [CrossRef]
- van Berlo, J.H.; Molkentin, J.D. An emerging consensus on cardiac regeneration. Nat. Med. 2014, 20, 1386–1393. [Google Scholar] [CrossRef]
- Chen, Z.; Zhu, W.; Bender, I.; Gong, W.; Kwak, I.Y.; Yellamilli, A.; Hodges, T.J.; Nemoto, N.; Zhang, J.; Garry, D.J.; et al. Pathologic Stimulus Determines Lineage Commitment of Cardiac C-kit(+) Cells. Circulation 2017, 136, 2359–2372. [Google Scholar] [CrossRef] [PubMed]
- Vagnozzi, R.J.; Sargent, M.A.; Lin, S.J.; Palpant, N.J.; Murry, C.E.; Molkentin, J.D. Genetic Lineage Tracing of Sca-1(+) Cells Reveals Endothelial but Not Myogenic Contribution to the Murine Heart. Circulation 2018, 138, 2931–2939. [Google Scholar] [CrossRef]
- Cahill, T.J.; Choudhury, R.P.; Riley, P.R. Heart regeneration and repair after myocardial infarction: Translational opportunities for novel therapeutics. Nat. Rev. Drug Discov. 2017, 16, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Gallet, R.; Dawkins, J.; Valle, J.; Simsolo, E.; de Couto, G.; Middleton, R.; Tseliou, E.; Luthringer, D.; Kreke, M.; Smith, R.R.; et al. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur. Heart. J. 2017, 38, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Mishra, R.; Bigham, G.E.; Wehman, B.; Khan, M.M.; Xu, H.; Saha, P.; Goo, Y.A.; Datla, S.R.; Chen, L.; et al. A Deep Proteome Analysis Identifies the Complete Secretome as the Functional Unit of Human Cardiac Progenitor Cells. Circ. Res. 2017, 120, 816–834. [Google Scholar] [CrossRef] [PubMed]
- Timmers, L.; Lim, S.K.; Arslan, F.; Armstrong, J.S.; Hoefer, I.E.; Doevendans, P.A.; Piek, J.J.; El Oakley, R.M.; Choo, A.; Lee, C.N.; et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2007, 1, 129–137. [Google Scholar] [CrossRef]
- Uhlen, M.; Tegel, H.; Sivertsson, Å.; Kuo, C.-C.; Gutierrez, J.M.; Lewis, N.E.; Forsström, B.; Dannemeyer, M.; Fagerberg, L.; Malm, M.; et al. The human secretome – the proteins secreted from human cells. bioRxiv 2018. [Google Scholar] [CrossRef]
- Wang, S.; Li, Y.; Jiang, C.; Tian, H. Fibroblast growth factor 9 subfamily and the heart. Appl. Microbiol. Biotechnol. 2017. [Google Scholar] [CrossRef]
- Drowley, L.; Koonce, C.; Peel, S.; Jonebring, A.; Plowright, A.T.; Kattman, S.J.; Andersson, H.; Anson, B.; Swanson, B.J.; Wang, Q.D.; et al. Human Induced Pluripotent Stem Cell-Derived Cardiac Progenitor Cells in Phenotypic Screening: A Transforming Growth Factor-beta Type 1 Receptor Kinase Inhibitor Induces Efficient Cardiac Differentiation. Stem Cells Transl. Med. 2016, 5, 164–174. [Google Scholar] [CrossRef]
- Paunovic, A.I.; Drowley, L.; Nordqvist, A.; Ericson, E.; Mouchet, E.; Jonebring, A.; Gronberg, G.; Kvist, A.J.; Engkvist, O.; Brown, M.R.; et al. Phenotypic Screen for Cardiac Regeneration Identifies Molecules with Differential Activity in Human Epicardium-Derived Cells versus Cardiac Fibroblasts. ACS Chem. Biol. 2017, 12, 132–141. [Google Scholar] [CrossRef]
- Zhang, X.; Ibrahimi, O.A.; Olsen, S.K.; Umemori, H.; Mohammadi, M.; Ornitz, D.M. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J. Biol. Chem. 2006, 281, 15694–15700. [Google Scholar] [CrossRef] [PubMed]
- Salanti, A.; Clausen, T.M.; Agerbaek, M.O.; Al Nakouzi, N.; Dahlback, M.; Oo, H.Z.; Lee, S.; Gustavsson, T.; Rich, J.R.; Hedberg, B.J.; et al. Targeting Human Cancer by a Glycosaminoglycan Binding Malaria Protein. Cancer Cell 2015, 28, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Drowley, L.; McPheat, J.; Nordqvist, A.; Peel, S.; Karlsson, U.; Martinsson, S.; Mullers, E.; Dellsen, A.; Knight, S.; Barrett, I.; et al. Discovery of Retinoic Acid Receptor Agonists as Proliferators of Cardiac Progenitor Cells Through a Phenotypic Screening Approach. Stem Cells Transl. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tian, L.; Shen, M.; Tu, C.; Wu, H.; Gu, M.; Paik, D.T.; Wu, J.C. Generation of Quiescent Cardiac Fibroblasts from Human Induced Pluripotent Stem Cells for in Vitro Modeling of Cardiac Fibrosis. Circ. Res. 2019, 125, 552–566. [Google Scholar] [CrossRef]
- Miyake, A.; Konishi, M.; Martin, F.H.; Hernday, N.A.; Ozaki, K.; Yamamoto, S.; Mikami, T.; Arakawa, T.; Itoh, N. Structure and expression of a novel member, FGF-16, on the fibroblast growth factor family. Biochem. Biophys. Res. Commun. 1998, 243, 148–152. [Google Scholar] [CrossRef]
- Miyamoto, M.; Naruo, K.; Seko, C.; Matsumoto, S.; Kondo, T.; Kurokawa, T. Molecular cloning of a novel cytokine cDNA encoding the ninth member of the fibroblast growth factor family, which has a unique secretion property. Mol. Cell. Biol. 1993, 13, 4251–4259. [Google Scholar] [CrossRef]
- Fon Tacer, K.; Bookout, A.L.; Ding, X.; Kurosu, H.; John, G.B.; Wang, L.; Goetz, R.; Mohammadi, M.; Kuro-o, M.; Mangelsdorf, D.J.; et al. Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol. Endocrinol. 2010, 24, 2050–2064. [Google Scholar] [CrossRef]
- Hotta, Y.; Sasaki, S.; Konishi, M.; Kinoshita, H.; Kuwahara, K.; Nakao, K.; Itoh, N. Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev. Dyn. 2008, 237, 2947–2954. [Google Scholar] [CrossRef]
- Yu, W.; Huang, X.; Tian, X.; Zhang, H.; He, L.; Wang, Y.; Nie, Y.; Hu, S.; Lin, Z.; Zhou, B.; et al. GATA4 regulates Fgf16 to promote heart repair after injury. Development 2016, 143, 936–949. [Google Scholar] [CrossRef]
- Sontag, D.P.; Wang, J.; Kardami, E.; Cattini, P.A. FGF-2 and FGF-16 protect isolated perfused mouse hearts from acute doxorubicin-induced contractile dysfunction. Cardiovasc. Toxicol. 2013, 13, 244–253. [Google Scholar] [CrossRef]
- Laurell, T.; Nilsson, D.; Hofmeister, W.; Lindstrand, A.; Ahituv, N.; Vandermeer, J.; Amilon, A.; Anneren, G.; Arner, M.; Pettersson, M.; et al. Identification of three novel FGF16 mutations in X-linked recessive fusion of the fourth and fifth metacarpals and possible correlation with heart disease. Mol. Genet. Genomic Med. 2014, 2, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Uccelli, A.; Wolff, T.; Valente, P.; Di Maggio, N.; Pellegrino, M.; Gurke, L.; Banfi, A.; Gianni-Barrera, R. Vascular endothelial growth factor biology for regenerative angiogenesis. Swiss Med. Wkly 2019, 149, w20011. [Google Scholar] [CrossRef] [PubMed]
- Zangi, L.; Lui, K.O.; von Gise, A.; Ma, Q.; Ebina, W.; Ptaszek, L.M.; Spater, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groppe, J.; Greenwald, J.; Wiater, E.; Rodriguez-Leon, J.; Economides, A.N.; Kwiatkowski, W.; Affolter, M.; Vale, W.W.; Izpisua Belmonte, J.C.; Choe, S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature 2002, 420, 636–642. [Google Scholar] [CrossRef]
- Kattman, S.J.; Witty, A.D.; Gagliardi, M.; Dubois, N.C.; Niapour, M.; Hotta, A.; Ellis, J.; Keller, G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 2011, 8, 228–240. [Google Scholar] [CrossRef] [Green Version]
- Boles, N.C.; Hirsch, S.E.; Le, S.; Corneo, B.; Najm, F.; Minotti, A.P.; Wang, Q.; Lotz, S.; Tesar, P.J.; Fasano, C.A. NPTX1 regulates neural lineage specification from human pluripotent stem cells. Cell Rep. 2014, 6, 724–736. [Google Scholar] [CrossRef] [Green Version]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [Green Version]
- Viklund, H.; Bernsel, A.; Skwark, M.; Elofsson, A. SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics 2008, 24, 2928–2929. [Google Scholar] [CrossRef] [Green Version]
- Fagerberg, L.; Jonasson, K.; von Heijne, G.; Uhlen, M.; Berglund, L. Prediction of the human membrane proteome. Proteomics 2010, 10, 1141–1149. [Google Scholar] [CrossRef]
- Brown, K.J.; Seol, H.; Pillai, D.K.; Sankoorikal, B.J.; Formolo, C.A.; Mac, J.; Edwards, N.J.; Rose, M.C.; Hathout, Y. The human secretome atlas initiative: Implications in health and disease conditions. Biochim. Biophys. Acta 2013, 1834, 2454–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, E.M.; van Oorschot, A.A.; Hogers, B.; van der Graaf, L.M.; Doevendans, P.A.; Poelmann, R.E.; Atsma, D.E.; Gittenberger-de Groot, A.C.; Goumans, M.J. A new direction for cardiac regeneration therapy: Application of synergistically acting epicardium-derived cells and cardiomyocyte progenitor cells. Circ. Heart Fail. 2009, 2, 643–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stastna, M.; Van Eyk, J.E. Investigating the secretome: Lessons about the cells that comprise the heart. Circ. Cardiovasc Genet. 2012, 5, o8–o18. [Google Scholar] [CrossRef] [PubMed]
- Smart, N.; Dube, K.N.; Riley, P.R. Epicardial progenitor cells in cardiac regeneration and neovascularisation. Vascul. Pharmacol. 2013, 58, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Aurora, A.B.; Porrello, E.R.; Tan, W.; Mahmoud, A.I.; Hill, J.A.; Bassel-Duby, R.; Sadek, H.A.; Olson, E.N. Macrophages are required for neonatal heart regeneration. J. Clin. Invest. 2014, 124, 1382–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lien, C.L.; Schebesta, M.; Makino, S.; Weber, G.J.; Keating, M.T. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006, 4, e260. [Google Scholar] [CrossRef]
- Daramola, O.; Stevenson, J.; Dean, G.; Hatton, D.; Pettman, G.; Holmes, W.; Field, R. A high-yielding CHO transient system: Coexpression of genes encoding EBNA-1 and GS enhances transient protein expression. Biotechnol. Prog. 2014, 30, 132–141. [Google Scholar] [CrossRef]
- Jansson, A.M.; Csiszar, A.; Maier, J.; Nystrom, A.C.; Ax, E.; Johansson, P.; Schiavone, L.H. The interleukin-like epithelial-mesenchymal transition inducer ILEI exhibits a non-interleukin-like fold and is active as a domain-swapped dimer. J. Biol. Chem. 2017, 292, 15501–15511. [Google Scholar] [CrossRef] [Green Version]
- Noseda, M.; Harada, M.; McSweeney, S.; Leja, T.; Belian, E.; Stuckey, D.J.; Abreu Paiva, M.S.; Habib, J.; Macaulay, I.; de Smith, A.J.; et al. PDGFRalpha demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat. Commun. 2015, 6, 6930. [Google Scholar] [CrossRef] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jennbacken, K.; Wågberg, F.; Karlsson, U.; Eriksson, J.; Magnusson, L.; Chimienti, M.; Ricchiuto, P.; Bernström, J.; Ding, M.; Ross-Thriepland, D.; et al. Phenotypic Screen with the Human Secretome Identifies FGF16 as Inducing Proliferation of iPSC-Derived Cardiac Progenitor Cells. Int. J. Mol. Sci. 2019, 20, 6037. https://doi.org/10.3390/ijms20236037
Jennbacken K, Wågberg F, Karlsson U, Eriksson J, Magnusson L, Chimienti M, Ricchiuto P, Bernström J, Ding M, Ross-Thriepland D, et al. Phenotypic Screen with the Human Secretome Identifies FGF16 as Inducing Proliferation of iPSC-Derived Cardiac Progenitor Cells. International Journal of Molecular Sciences. 2019; 20(23):6037. https://doi.org/10.3390/ijms20236037
Chicago/Turabian StyleJennbacken, Karin, Fredrik Wågberg, Ulla Karlsson, Jerry Eriksson, Lisa Magnusson, Marjorie Chimienti, Piero Ricchiuto, Jenny Bernström, Mei Ding, Douglas Ross-Thriepland, and et al. 2019. "Phenotypic Screen with the Human Secretome Identifies FGF16 as Inducing Proliferation of iPSC-Derived Cardiac Progenitor Cells" International Journal of Molecular Sciences 20, no. 23: 6037. https://doi.org/10.3390/ijms20236037
APA StyleJennbacken, K., Wågberg, F., Karlsson, U., Eriksson, J., Magnusson, L., Chimienti, M., Ricchiuto, P., Bernström, J., Ding, M., Ross-Thriepland, D., Xue, Y., Peiris, D., Aastrup, T., Tegel, H., Hober, S., Sivertsson, Å., Uhlén, M., Strömstedt, P.-E., Davies, R., & Holmberg Schiavone, L. (2019). Phenotypic Screen with the Human Secretome Identifies FGF16 as Inducing Proliferation of iPSC-Derived Cardiac Progenitor Cells. International Journal of Molecular Sciences, 20(23), 6037. https://doi.org/10.3390/ijms20236037