The Role of C-X-C Chemokine Receptor Type 4 (CXCR4) in Cell Adherence and Spheroid Formation of Human Ewing’s Sarcoma Cells under Simulated Microgravity
Abstract
:1. Introduction
2. Results
2.1. Light Microscopy
2.2. Trypan Blue Staining
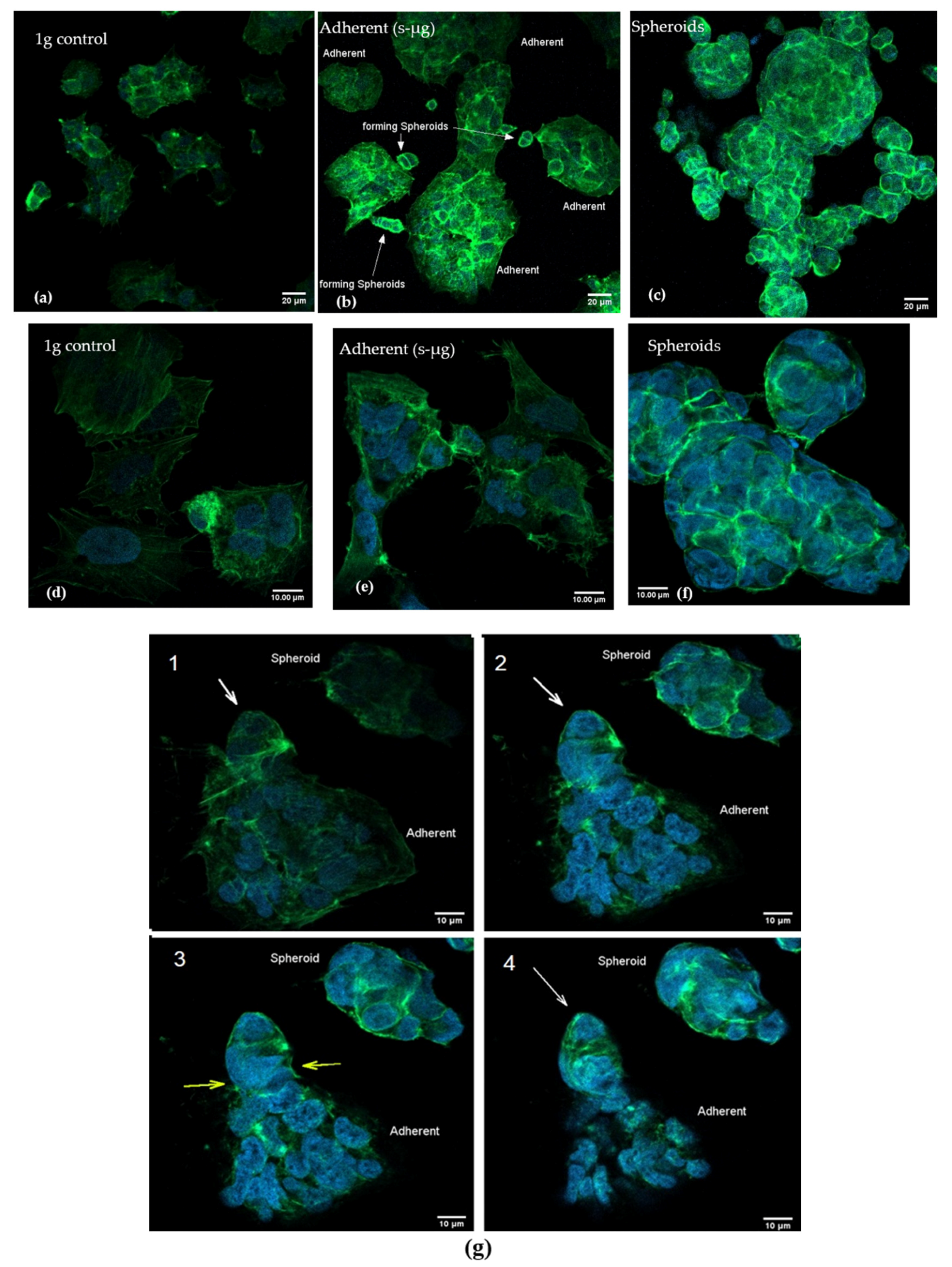
2.3. Confocal Microscopy after Actin Staining
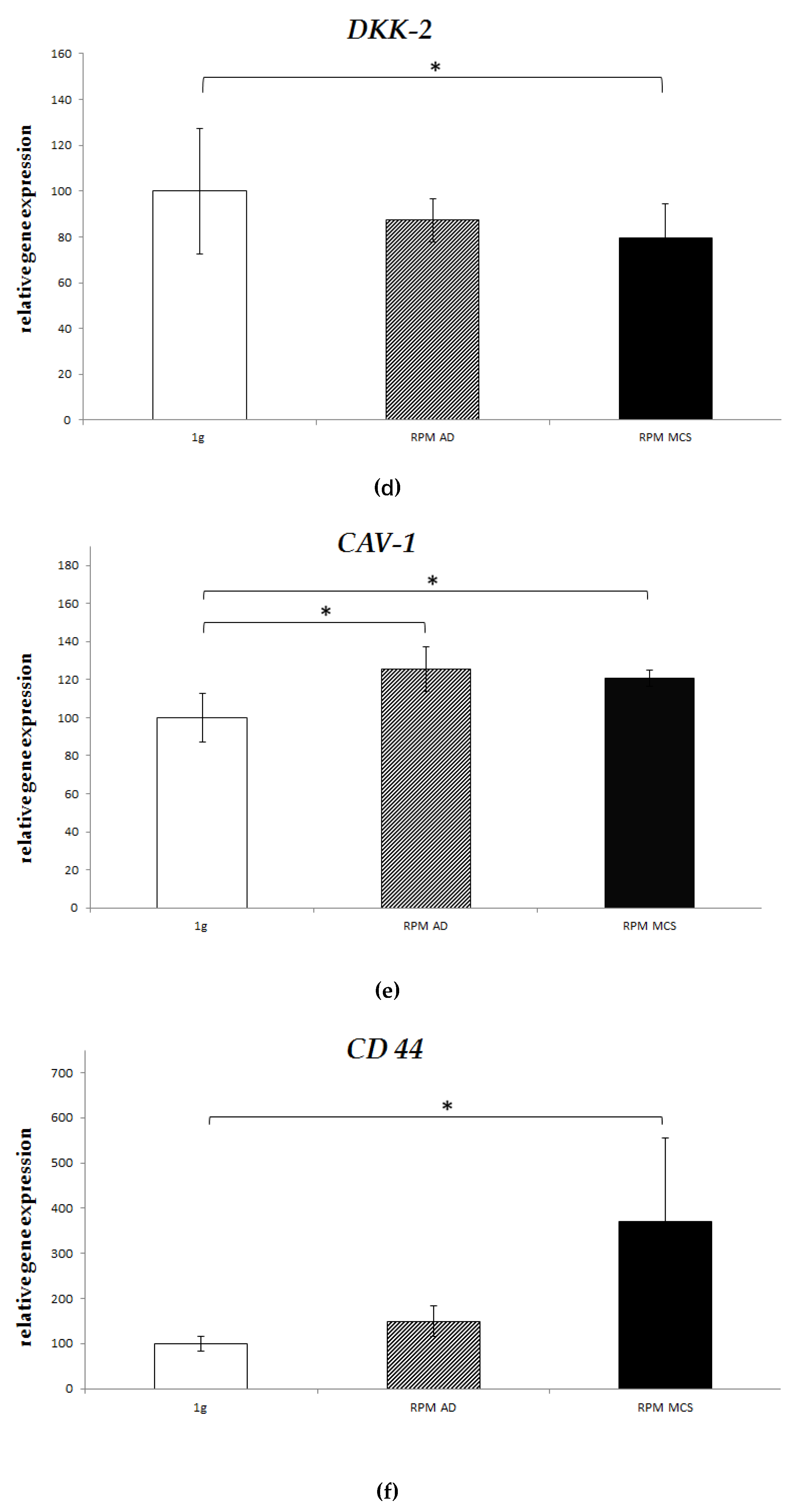
2.4. Real-Time PCR
2.5. Western-Blot
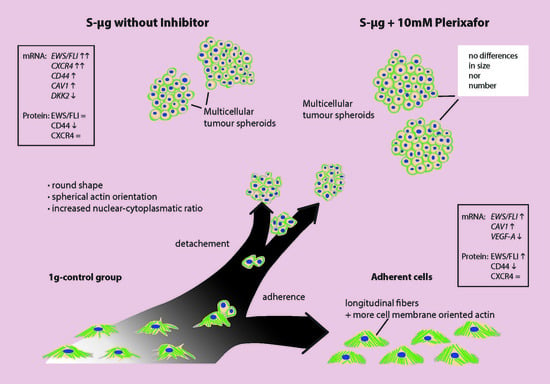
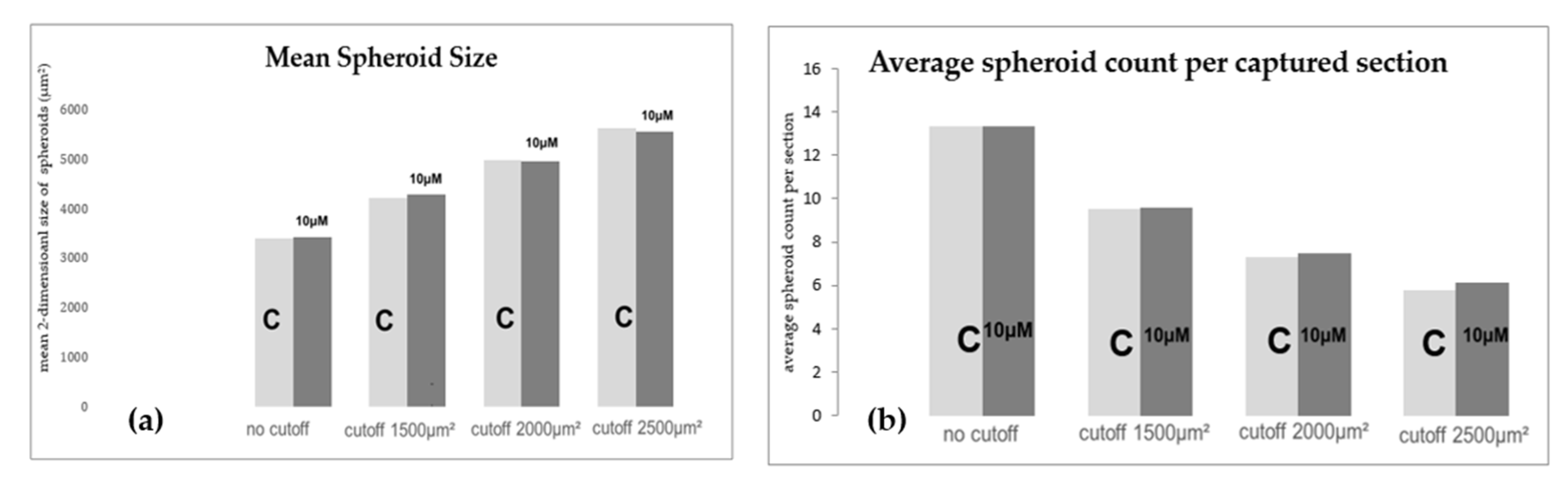
2.6. Morphologic and Quantitative Analysis of CXCR4-Inhibition with Plerixafor (AMD 3100) after 24 h RPM-Exposure
2.7. Protein-to-Protein Interaction Network via STRING Analysis
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Random Positioning Machine
4.3. Phase Contrast Microscopy
4.4. Trypan Blue Vitality Staining
4.5. Cytoskeleton Staining
4.6. Confocal Microscopy
4.7. RNA Isolation
4.8. Reverse Transcription
4.9. qRT-PCR
4.10. Western Blots
4.11. Morphologic and Quantitative Analysis of CXCR4-Inhibition with Plerixafor (AMD 3100) after 24 h RPM-Exposure
4.12. Protein-to-Protein Interaction Network via STRING Analysis
4.13. Statistical Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 1g | gravitational force equivalent of 1 |
| 2D | two-dimensional |
| 3D | three-dimensional |
| DAPI | 4′,6-diamidine-2-phenylindol |
| DNA | Deoxyribonucleic acid |
| cDNA | complementary DNA |
| DPBS | Dulbecco’s phosphate-buffered saline |
| ECM | Extracellular matrix |
| e.g., | Exempli gratia |
| ES | Ewing’s sarcoma |
| FCS | fetal calf serum |
| kD | Kilodalton |
| MCS | Multicellular spheroids |
| MW | molecular weight |
| PBS | phosphate-buffered saline |
| r-µg | Real microgravity |
| s-µg | Simulated microgravity |
| RNA | ribonucleic acid |
| RBP | RNA-binding protein |
| mRNA | messenger ribonucleic acid |
| miRNA | micro RNA |
| RPM | Random Positioning Machine |
| rpm | rounds per min |
| UPR | unfolded protein response |
References
- Rodríguez-Galindo, C.; Liu, T.; Krasin, M.J.; Wu, J.; Billups, C.A.; Daw, N.C.; Spunt, S.L.; Rao, B.N.; Santana, V.M.; Navid, F. Analysis of prognostic factors in ewing sarcoma family of tumors: Review of St. Jude Children’s Research Hospital studies. Cancer 2007, 110, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Ries, L.A.G.; Smith, M.A.; Gurney, J.G.; Linet, M.; Tamra, T.; Young, J.L.; Bunin, G.R. Cancer Incidence and Survival among Children SEER Program; National Institutes of Health: Bethesda, MD, USA, 1999; p. 192. [Google Scholar]
- Khanna, N.; Pandey, A.; Bajpai, J. Metastatic Ewing’s Sarcoma: Revisiting the “evidence on the fence”. Indian J. Med. Paediatr. Oncol. Off. J. Indian Soc. Med. Paediatr. Oncol. 2017, 38, 173–181. [Google Scholar]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; Le Deley, M.-C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current management and future approaches through collaboration. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef] [PubMed]
- Nesbit, M.E.; Gehan, E.A.; Burgert, E.O.; Vietti, T.J.; Cangir, A.; Tefft, M.; Evans, R.; Thomas, P.; Askin, F.B.; Kissane, J.M. Multimodal therapy for the management of primary, nonmetastatic Ewing’s sarcoma of bone: A long-term follow-up of the First Intergroup study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1990, 8, 1664–1674. [Google Scholar] [CrossRef] [PubMed]
- Pishas, K.I.; Lessnick, S.L. Recent advances in targeted therapy for Ewing sarcoma. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309. [Google Scholar] [CrossRef]
- Ghosh, S.; Spagnoli, G.C.; Martin, I.; Ploegert, S.; Demougin, P.; Heberer, M.; Reschner, A. Three-dimensional culture of melanoma cells profoundly affects gene expression profile: A high density oligonucleotide array study. J. Cell. Physiol. 2005, 204, 522–531. [Google Scholar] [CrossRef]
- Hickman, J.A.; Graeser, R.; Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; van der Kuip, H. Three-dimensional models of cancer for pharmacology and cancer cell biology: Capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef]
- Santini, M. Apoptosis, cell adhesion and the extracellular matrix in the three-dimensional growth of multicellular tumor spheroids. Crit. Rev. Oncol. Hematol. 2000, 36, 75–87. [Google Scholar] [CrossRef]
- Lawlor, E.R.; Scheel, C.; Irving, J.; Sorensen, P.H.B. Anchorage-independent multi-cellular spheroids as an in vitro model of growth signaling in Ewing tumors. Oncogene 2002, 21, 307. [Google Scholar] [CrossRef] [PubMed]
- Norsk, P. Blood pressure regulation IV: Adaptive responses to weightlessness. Eur. J. Appl. Physiol. 2014, 114, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Abrams, S.A.; Davis-Street, J.E.; Heer, M.; O′Brien, K.O.; Wastney, M.E.; Zwart, S.R. Fifty years of human space travel: Implications for bone and calcium research. Annu. Rev. Nutr. 2014, 34, 377–400. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, A.; Grimm, D.; Sahana, J.; Bauer, J.; Krüger, M.; Infanger, M.; Magnusson, N.E. Key Proteins Involved in Spheroid Formation and Angiogenesis in Endothelial Cells After Long-Term Exposure to Simulated Microgravity. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 45, 429–445. [Google Scholar] [CrossRef]
- Ma, X.; Sickmann, A.; Pietsch, J.; Wildgruber, R.; Weber, G.; Infanger, M.; Bauer, J.; Grimm, D. Proteomic differences between microvascular endothelial cells and the EA.hy926 cell line forming three-dimensional structures. Proteomics 2014, 14, 689–698. [Google Scholar] [CrossRef]
- Pietsch, J.; Gass, S.; Nebuloni, S.; Echegoyen, D.; Riwaldt, S.; Baake, C.; Bauer, J.; Corydon, T.J.; Egli, M.; Infanger, M.; et al. Three-dimensional growth of human endothelial cells in an automated cell culture experiment container during the SpaceX CRS-8 ISS space mission—The SPHEROIDS project. Biomaterials 2017, 124, 126–156. [Google Scholar] [CrossRef]
- Grimm, D.; Bauer, J.; Ulbrich, C.; Westphal, K.; Wehland, M.; Infanger, M.; Aleshcheva, G.; Pietsch, J.; Ghardi, M.; Beck, M.; et al. Different responsiveness of endothelial cells to vascular endothelial growth factor and basic fibroblast growth factor added to culture media under gravity and simulated microgravity. Tissue Eng. Part A 2010, 16, 1559–1573. [Google Scholar] [CrossRef]
- Ulbrich, C.; Westphal, K.; Baatout, S.; Wehland, M.; Bauer, J.; Flick, B.; Infanger, M.; Kreutz, R.; Vadrucci, S.; Egli, M.; et al. Effects of basic fibroblast growth factor on endothelial cells under conditions of simulated microgravity. J. Cell. Biochem. 2008, 104, 1324–1341. [Google Scholar] [CrossRef]
- Kraus, A.; Luetzenberg, R.; Abuagela, N.; Hollenberg, S.; Infanger, M. Spheroid formation and modulation of tenocyte-specific gene expression under simulated microgravity. Muscles Ligaments Tendons J. 2017, 7, 411–417. [Google Scholar] [CrossRef]
- Wehland, M.; Aleshcheva, G.; Schulz, H.; Saar, K.; Hübner, N.; Hemmersbach, R.; Braun, M.; Ma, X.; Frett, T.; Warnke, E.; et al. Differential gene expression of human chondrocytes cultured under short-term altered gravity conditions during parabolic flight maneuvers. Cell Commun. Signal. CCS 2015, 13, 18. [Google Scholar] [CrossRef]
- Bauer, J.; Kopp, S.; Schlagberger, E.M.; Grosse, J.; Sahana, J.; Riwaldt, S.; Wehland, M.; Luetzenberg, R.; Infanger, M.; Grimm, D. Proteome analysis of human follicular thyroid cancer cells exposed to the random positioning machine. Int. J. Mol. Sci. 2017, 18, 546. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Wehland, M.; Infanger, M.; Grimm, D.; Gombocz, E. Semantic analysis of posttranslational modification of proteins accumulated in thyroid cancer cells exposed to simulated microgravity. Int. J. Mol. Sci. 2018, 19, 2257. [Google Scholar] [CrossRef] [PubMed]
- Warnke, E.; Pietsch, J.; Wehland, M.; Bauer, J.; Infanger, M.; Görög, M.; Hemmersbach, R.; Braun, M.; Ma, X.; Sahana, J.; et al. Spheroid formation of human thyroid cancer cells under simulated microgravity: A possible role of CTGF and CAV1. Cell Commun. Signal. CCS 2014, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Kopp, S.; Slumstrup, L.; Corydon, T.J.; Sahana, J.; Aleshcheva, G.; Islam, T.; Magnusson, N.E.; Wehland, M.; Bauer, J.; Infanger, M.; et al. Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the Random Positioning Machine. Sci. Rep. 2016, 6, 26887. [Google Scholar] [CrossRef] [PubMed]
- Kopp, S.; Krüger, M.; Feldmann, S.; Oltmann, H.; Schütte, A.; Schmitz, B.; Bauer, J.; Schulz, H.; Saar, K.; Huebner, N.; et al. Thyroid cancer cells in space during the TEXUS-53 sounding rocket mission—The THYROID Project. Sci. Rep. 2018, 8, 10355. [Google Scholar] [CrossRef]
- Pietsch, J.; Ma, X.; Wehland, M.; Aleshcheva, G.; Schwarzwälder, A.; Segerer, J.; Birlem, M.; Horn, A.; Bauer, J.; Infanger, M.; et al. Spheroid formation of human thyroid cancer cells in an automated culturing system during the Shenzhou-8 Space mission. Biomaterials 2013, 34, 7694–7705. [Google Scholar] [CrossRef]
- Sahebi, R.; Aghaei, M.; Halvaei, S.; Alizadeh, A. The role of microgravity in cancer: A dual-edge sword. Multidiscip. Cancer Investig. 2017, 1, 1–2. [Google Scholar] [CrossRef]
- Masiello, M.G.; Cucina, A.; Proietti, S.; Palombo, A.; Coluccia, P.; D′Anselmi, F.; Dinicola, S.; Pasqualato, A.; Morini, V.; Bizzarri, M. Phenotypic switch induced by simulated microgravity on MDA-MB-231 breast cancer cells. BioMed Res. Int. 2014, 2014. [Google Scholar] [CrossRef]
- Wuest, S.L.; Richard, S.; Kopp, S.; Grimm, D.; Egli, M. Simulated microgravity: Critical review on the use of random positioning machines for mammalian cell culture. BioMed Res. Int. 2015, 2015, 971474. [Google Scholar] [CrossRef]
- Borst, A.G.; van Loon, J.J.W.A. Technology and Developments for the Random Positioning Machine, RPM. Microgravity Sci. Technol. 2008, 21, 287. [Google Scholar] [CrossRef]
- Pietsch, J.; Sickmann, A.; Weber, G.; Bauer, J.; Egli, M.; Wildgruber, R.; Infanger, M.; Grimm, D. A proteomic approach to analysing spheroid formation of two human thyroid cell lines cultured on a random positioning machine. Proteomics 2011, 11, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Svejgaard, B.; Wehland, M.; Ma, X.; Kopp, S.; Sahana, J.; Warnke, E.; Aleshcheva, G.; Hemmersbach, R.; Hauslage, J.; Grosse, J.; et al. Common effects on cancer cells exerted by a random positioning machine and a 2D Clinostat. PloS ONE 2015, 10, e0135157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimm, D.; Wehland, M.; Pietsch, J.; Aleshcheva, G.; Wise, P.; van Loon, J.; Ulbrich, C.; Magnusson, N.E.; Infanger, M.; Bauer, J. Growing tissues in real and simulated microgravity: New methods for tissue engineering. Tissue Eng. Part B Rev. 2014, 20, 555–566. [Google Scholar] [CrossRef] [Green Version]
- Terzis, A.-J.; Thorsen, F.; Heese, O.; Visted, T.; Bjerkvig, R.; Dahl, O.; Arnold, H.; Gundersen, G. Proliferation, migration and invasion of human glioma cells exposed to paclitaxel (Taxol) in vitro. Br. J. Cancer 1997, 75, 1744–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankel, A.; Buckman, R.; Kerbel, R.S. Abrogation of taxol-induced G2-M arrest and apoptosis in human ovarian cancer cells grown as multicellular tumor spheroids. Cancer Res. 1997, 57, 2388–2393. [Google Scholar]
- Santini, M.T.; Rainaldi, G.; Indovina, P.L. Multicellular tumour spheroids in radiation biology. Int. J. Radiat. Biol. 1999, 75, 787–799. [Google Scholar] [CrossRef]
- Hamilton, G. Multicellular spheroids as an in vitro tumor model. Cancer Lett. 1998, 131, 29–34. [Google Scholar] [CrossRef]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [Green Version]
- Mehta, G.; Hsiao, A.Y.; Ingram, M.; Luker, G.D.; Takayama, S. Opportunities and Challenges for use of Tumor Spheroids as Models to Test Drug Delivery and Efficacy. J. Control. Release Off. J. Control. Release Soc. 2012, 164, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Groebe, K.; Mueller-Klieser, W. Distributions of oxygen, nutrient, and metabolic waste concentrations in multicellular spheroids and their dependence on spheroid parameters. Eur. Biophys. J. 1991, 19, 169–181. [Google Scholar] [CrossRef]
- Chatzinikolaidou, M. Cell spheroids: The new frontiers in in vitro models for cancer drug validation. Drug Discov. Today 2016, 21, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Kruger, M.; Melnik, D.; Kopp, S.; Buken, C.; Sahana, J.; Bauer, J.; Wehland, M.; Hemmersbach, R.; Corydon, T.J.; Infanger, M.; et al. Fighting thyroid cancer with microgravity research. Int. J. Mol. Sci. 2019, 20, 2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clejan, S.; O′Connor, K.C.; Cowger, N.L.; Cheles, M.K.; Haque, S.; Primavera, A.C. Effects of simulated microgravity on DU 145 human prostate carcinoma cells. Biotechnol. Bioeng. 1996, 50, 587–597. [Google Scholar] [CrossRef]
- Ma, X.; Pietsch, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Bauer, J.; Braun, M.; Schwarzwalder, A.; Segerer, J.; et al. Differential gene expression profile and altered cytokine secretion of thyroid cancer cells in space. FASEB J. 2014, 28, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Magaki, T.; Okazaki, T.; Kawahara, Y.; Manabe, T.; Yuge, L.; Kurisu, K. Effects of simulated microgravity on proliferation and chemosensitivity in malignant glioma cells. Neurosci. Lett. 2009, 463, 54–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, A.; Hoffman, L.M.; Welm, A.L.; Lessnick, S.L.; Beckerle, M.C. The EWS/FLI oncogene drives changes in cellular morphology, adhesion, and migration in ewing sarcoma. Genes Cancer 2012, 3, 102–116. [Google Scholar] [CrossRef] [Green Version]
- Wiles, E.T.; Bell, R.; Thomas, D.; Beckerle, M.; Lessnick, S.L. ZEB2 represses the epithelial phenotype and facilitates metastasis in ewing sarcoma. Genes Cancer 2013, 4, 486–500. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.X.; Zhou, C.H.; Zeng, H.; Zuo, D.Q.; Wang, Z.Y.; Yin, F.; Hua, Y.Q.; Cai, Z.D. The role of the CXCL12-CXCR4/CXCR7 axis in the progression and metastasis of bone sarcomas (Review). Int. J. Mol. Med. 2013, 32, 1239–1246. [Google Scholar] [CrossRef]
- Paulis, Y.W.; Huijbers, E.J.; van der Schaft, D.W.; Soetekouw, P.M.; Pauwels, P.; Tjan-Heijnen, V.C.; Griffioen, A.W. CD44 enhances tumor aggressiveness by promoting tumor cell plasticity. Oncotarget 2015, 6, 19634–19646. [Google Scholar] [CrossRef] [Green Version]
- Sainz-Jaspeado, M.; Lagares-Tena, L.; Lasheras, J.; Navid, F.; Rodriguez-Galindo, C.; Mateo-Lozano, S.; Notario, V.; Sanjuan, X.; Garcia Del Muro, X.; Fabra, A.; et al. Caveolin-1 modulates the ability of Ewing’s sarcoma to metastasize. Mol. Cancer Res. 2010, 8, 1489–1500. [Google Scholar] [CrossRef] [Green Version]
- Tirado, O.M.; Mateo-Lozano, S.; Villar, J.; Dettin, L.E.; Llort, A.; Gallego, S.; Ban, J.; Kovar, H.; Notario, V. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006, 66, 9937–9947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagares-Tena, L.; Garcia-Monclus, S.; Lopez-Alemany, R.; Almacellas-Rabaiget, O.; Huertas-Martinez, J.; Sainz-Jaspeado, M.; Mateo-Lozano, S.; Rodriguez-Galindo, C.; Rello-Varona, S.; Herrero-Martin, D.; et al. Caveolin-1 promotes Ewing sarcoma metastasis regulating MMP-9 expression through MAPK/ERK pathway. Oncotarget 2016, 7, 56889–56903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spisni, E.; Toni, M.; Strillacci, A.; Galleri, G.; Santi, S.; Griffoni, C.; Tomasi, V. Caveolae and caveolae constituents in mechanosensing: Effect of modeled microgravity on cultured human endothelial cells. Cell Biochem. Biophys. 2006, 46, 155–164. [Google Scholar] [CrossRef]
- Shi, F.; Zhao, T.Z.; Wang, Y.C.; Cao, X.S.; Yang, C.B.; Gao, Y.; Li, C.F.; Zhao, J.D.; Zhang, S.; Sun, X.Q. The impact of simulated weightlessness on endothelium-dependent angiogenesis and the role of caveolae/caveolin-1. Cell Physiol. Biochem. 2016, 38, 502–513. [Google Scholar] [CrossRef]
- Becker, J.L.; Souza, G.R. Using space-based investigations to inform cancer research on Earth. Nat. Rev. Cancer 2013, 13, 315–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Dietz, C.; Infanger, M.; Romswinkel, A.; Strube, F.; Kraus, A. Apoptosis induction and alteration of cell adherence in human lung cancer cells under simulated microgravity. Int. J. Mol. Sci. 2019, 20, 3601. [Google Scholar] [CrossRef] [Green Version]
- Bachir, A.I.; Horwitz, A.R.; Nelson, W.J.; Bianchini, J.M. Actin-based adhesion modules mediate cell interactions with the extracellular matrix and neighboring cells. Cold Spring Harbor Perspect. Biol. 2017, 9, 3601. [Google Scholar] [CrossRef] [Green Version]
- van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef]
- Vorselen, D.; Roos, W.H.; MacKintosh, F.C.; Wuite, G.J.; van Loon, J.J. The role of the cytoskeleton in sensing changes in gravity by nonspecialized cells. FASEB J. 2014, 28, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Corydon, T.J.; Kopp, S.; Wehland, M.; Braun, M.; Schutte, A.; Mayer, T.; Hulsing, T.; Oltmann, H.; Schmitz, B.; Hemmersbach, R.; et al. Alterations of the cytoskeleton in human cells in space proved by life-cell imaging. Sci. Rep. 2016, 6, 20043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi, K.; Fujiwara, S.; Mizuno, K. Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. J. Biochem. 2017, 161, 245–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riboh, J.; Chong, A.K.; Pham, H.; Longaker, M.; Jacobs, C.; Chang, J. Optimization of flexor tendon tissue engineering with a cyclic strain bioreactor. J. Hand Surg. 2008, 33, 1388–1396. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.; Lessnick, S.L. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011, 204, 351–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.; Owen, L.A.; Trem, D.J.; Wong, J.S.; Whangbo, J.S.; Golub, T.R.; Lessnick, S.L. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell 2006, 9, 405–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauschke, V.M.; Shafagh, R.Z.; Hendriks, D.F.G.; Ingelman-Sundberg, M. 3D Primary hepatocyte culture systems for analyses of liver diseases, drug metabolism, and toxicity: Emerging culture paradigms and applications. Biotechnol. J. 2019, 14, e1800347. [Google Scholar] [CrossRef] [Green Version]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [Green Version]
- Lamhamedi-Cherradi, S.E.; Santoro, M.; Ramammoorthy, V.; Menegaz, B.A.; Bartholomeusz, G.; Iles, L.R.; Amin, H.M.; Livingston, J.A.; Mikos, A.G.; Ludwig, J.A. 3D tissue-engineered model of Ewing’s sarcoma. Adv. Drug Deliver. Rev. 2014, 79–80, 155–171. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.; Lamhamedi-Cherradi, S.E.; Menegaz, B.A.; Ludwig, J.A.; Mikos, A.G. Flow perfusion effects on three-dimensional culture and drug sensitivity of Ewing sarcoma. Proc. Natl. Acad. Sci. USA 2015, 112, 10304–10309. [Google Scholar] [CrossRef] [Green Version]
- Konemann, S.; Malath, J.; Bolling, T.; Kolkmeyer, A.; Janke, K.; Riesenbeck, D.; Hesselmann, S.; Nguyen, T.P.; Diallo, R.; Vormoor, J.; et al. Changed adhesion molecule profile of Ewing tumor cell lines and xenografts under the influence of ionizing radiation. Anticancer Res. 2004, 24, 1637–1644. [Google Scholar]
- Hauer, K.; Calzada-Wack, J.; Steiger, K.; Grunewald, T.G.; Baumhoer, D.; Plehm, S.; Buch, T.; Prazeres da Costa, O.; Esposito, I.; Burdach, S.; et al. DKK2 mediates osteolysis, invasiveness, and metastatic spread in Ewing sarcoma. Cancer Res. 2013, 73, 967–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scala, S. Molecular Pathways: Targeting the CXCR4-CXCL12 Axis—Untapped potential in the tumor Microenvironment. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 4278–4285. [Google Scholar] [CrossRef] [PubMed]
- Aryee, D.N.; Niedan, S.; Kauer, M.; Schwentner, R.; Bennani-Baiti, I.M.; Ban, J.; Muehlbacher, K.; Kreppel, M.; Walker, R.L.; Meltzer, P.; et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing’s sarcoma cells in vitro. Cancer Res. 2010, 70, 4015–4023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krook, M.A.; Nicholls, L.A.; Scannell, C.A.; Chugh, R.; Thomas, D.G.; Lawlor, E.R. Stress-induced CXCR4 promotes migration and invasion of ewing sarcoma. Mol. Cancer Res. MCR 2014, 12, 953–964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burger, J.A.; Kipps, T.J. CXCR4: A key receptor in the crosstalk between tumor cells and their microenvironment. Blood 2006, 107, 1761–1767. [Google Scholar] [CrossRef]
- Berghuis, D.; Schilham, M.W.; Santos, S.J.; Savola, S.; Knowles, H.J.; Dirksen, U.; Schaefer, K.L.; Vakkila, J.; Hogendoorn, P.C.; Lankester, A.C. The CXCR4-CXCL12 axis in Ewing sarcoma: Promotion of tumor growth rather than metastatic disease. Clin. Sarcoma Res. 2012, 2, 24. [Google Scholar] [CrossRef] [Green Version]
- Gassenmaier, M.; Chen, D.; Buchner, A.; Henkel, L.; Schiemann, M.; Mack, B.; Schendel, D.J.; Zimmermann, W.; Pohla, H. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells 2013, 31, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.J.; Rho, J.K.; Kim, Y.M.; Jung, J.E.; Jin, Y.B.; Ko, Y.G.; Lee, J.S.; Lee, S.J.; Lee, J.C.; Park, M.J. Upregulation of CXCR4 is functionally crucial for maintenance of stemness in drug-resistant non-small cell lung cancer cells. Oncogene 2013, 32, 209–221. [Google Scholar] [CrossRef]
- Feger, B.J.; Thompson, J.W.; Dubois, L.G.; Kommaddi, R.P.; Foster, M.W.; Mishra, R.; Shenoy, S.K.; Shibata, Y.; Kidane, Y.H.; Moseley, M.A.; et al. Microgravity induces proteomics changes involved in endoplasmic reticulum stress and mitochondrial protection. Sci. Rep. 2016, 6, 34091. [Google Scholar] [CrossRef] [Green Version]
- Nickerson, C.A.; Ott, C.M.; Mister, S.J.; Morrow, B.J.; Burns-Keliher, L.; Pierson, D.L. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2000, 68, 3147–3152. [Google Scholar] [CrossRef] [Green Version]
- Higashibata, A.; Hashizume, T.; Nemoto, K.; Higashitani, N.; Etheridge, T.; Mori, C.; Harada, S.; Sugimoto, T.; Szewczyk, N.J.; Baba, S.A.; et al. Microgravity elicits reproducible alterations in cytoskeletal and metabolic gene and protein expression in space-flown Caenorhabditis elegans. NPJ Microgravity 2016, 2, 15022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikemoto, M.; Nikawa, T.; Takeda, S.; Watanabe, C.; Kitano, T.; Baldwin, K.M.; Izumi, R.; Nonaka, I.; Towatari, T.; Teshima, S.; et al. Space shuttle flight (STS-90) enhances degradation of rat myosin heavy chain in association with activation of ubiquitin-proteasome pathway. FASEB J. 2001, 15, 1279–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirasaka, K.; Nikawa, T.; Yuge, L.; Ishihara, I.; Higashibata, A.; Ishioka, N.; Okubo, A.; Miyashita, T.; Suzue, N.; Ogawa, T.; et al. Clinorotation prevents differentiation of rat myoblastic L6 cells in association with reduced NF-kappa B signaling. Biochim. Biophys. Acta 2005, 1743, 130–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazarian, V.A.; Rapoport, E.A.; Goncharova, L.A.; Bulycheva, S. [Effect of prolonged weightlessness on protein metabolism in rat red and white skeletal muscles]. Kosmicheskaia Biol. Aviakosmicheskaia Med. 1977, 11, 19–23. [Google Scholar]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Siomi, H.; Siomi, M.C. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell 2010, 38, 323–332. [Google Scholar] [CrossRef]
- Agami, R. microRNAs, RNA binding proteins and cancer. Eur. J. Clin. Investig. 2010, 40, 370–374. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Mangala, L.S.; Zhang, Y.; He, Z.; Emami, K.; Ramesh, G.T.; Story, M.; Rohde, L.H.; Wu, H. Effects of simulated microgravity on expression profile of microRNA in human lymphoblastoid cells. J. Biol. Chem. 2011, 286, 32483–32490. [Google Scholar] [CrossRef] [Green Version]
- Girardi, C.; De Pitta, C.; Casara, S.; Calura, E.; Romualdi, C.; Celotti, L.; Mognato, M. Integration analysis of microRNA and mRNA expression profiles in human peripheral blood lymphocytes cultured in modeled microgravity. Biomed. Res. Int. 2014, 2014, 296747. [Google Scholar] [CrossRef] [Green Version]
- Teodori, L.; Costa, A.; Campanella, L.; Albertini, M.C. Skeletal muscle atrophy in simulated microgravity might be triggered by immune-related microRNAs. Front. Physiol. 2018, 9, 1926. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Brodie, E.L.; Matin, A. Role and regulation of sigma S in general resistance conferred by low-shear simulated microgravity in Escherichia coli. J. Bacteriol. 2004, 186, 8207–8212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Clercq, E. Mozobil(R) (Plerixafor, AMD3100), 10 years after its approval by the US Food and Drug Administration. Antivir. Chem. Chemother. 2019, 27, 2040206619829382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudary, N.; Pintilie, M.; Jelveh, S.; Lindsay, P.; Hill, R.P.; Milosevic, M. Plerixafor improves primary tumor response and reduces metastases in cervical cancer treated with radio-chemotherapy. Clin. Cancer Res. 2017, 23, 1242–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.; Xu, A.; Zhao, T.; Zhao, Q.; Zhang, J.; Fan, C.; Deng, Y.; Freywald, A.; Genth, H.; Xiang, J. Simulated microgravity inhibits cell focal adhesions leading to reduced melanoma cell proliferation and metastasis via FAK/RhoA-regulated mTORC1 and AMPK pathways. Sci. Rep. 2018, 8, 3769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benavides Damm, T.; Walther, I.; Wuest, S.L.; Sekler, J.; Egli, M. Cell cultivation under different gravitational loads using a novel random positioning incubator. Biotechnol. Bioeng. 2014, 111, 1180–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzenberg, M.; Pippia, P.; Meloni, M.A.; Cossu, G.; Cogoli-Greuter, M.; Cogoli, A. Signal transduction in T lymphocytes—A comparison of the data from space, the free fall machine and the random positioning machine. Adv. Space Res. Off. J. Comm. Space Res. 1999, 24, 793–800. [Google Scholar] [CrossRef]
- Herranz, R.; Benguria, A.; Lavan, D.A.; Lopez-Vidriero, I.; Gasset, G.; Javier Medina, F.; van Loon, J.J.; Marco, R. Spaceflight-related suboptimal conditions can accentuate the altered gravity response of Drosophila transcriptome. Mol. Ecol. 2010, 19, 4255–4264. [Google Scholar] [CrossRef]
- Pietsch, J.; Sickmann, A.; Weber, G.; Bauer, J.; Egli, M.; Wildgruber, R.; Infanger, M.; Grimm, D. Metabolic enzyme diversity in different human thyroid cell lines and their sensitivity to gravitational forces. Proteomics 2012, 12, 2539–2546. [Google Scholar] [CrossRef]
- Infanger, M.; Kossmehl, P.; Shakibaei, M.; Bauer, J.; Kossmehl-Zorn, S.; Cogoli, A.; Curcio, F.; Oksche, A.; Wehland, M.; Kreutz, R.; et al. Simulated weightlessness changes the cytoskeleton and extracellular matrix proteins in papillary thyroid carcinoma cells. Cell Tissue Res. 2006, 324, 267–277. [Google Scholar] [CrossRef]
- Erler, J.T.; Giaccia, A.J. Lysyl oxidase mediates hypoxic control of metastasis. Cancer Res. 2006, 66, 10238–10241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanceau, J.; Truchet, S.; Bauvois, B. Matrix metalloproteinase-9 silencing by RNA interference triggers the migratory-adhesive switch in Ewing’s sarcoma cells. J. Biol. Chem. 2003, 278, 36537–36546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Xu, C.; Ayello, J.; Dela Cruz, F.; Rosenblum, J.M.; Lessnick, S.L.; Cairo, M.S. Protein phosphatase 1 regulatory subunit 1A in ewing sarcoma tumorigenesis and metastasis. Oncogene 2018, 37, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Naranjo, A.; El-Naggar, A.; Wai, D.H.; Mistry, P.; Lazic, N.; Ayala, F.R.; da Cunha, I.W.; Rodriguez-Viciana, P.; Cheng, H.; Tavares Guerreiro Fregnani, J.H.; et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol. Med. 2013, 5, 1087–1102. [Google Scholar] [CrossRef]
- Kang, H.G.; Jenabi, J.M.; Zhang, J.; Keshelava, N.; Shimada, H.; May, W.A.; Ng, T.; Reynolds, C.P.; Triche, T.J.; Sorensen, P.H. E-cadherin cell-cell adhesion in ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007, 67, 3094–3105. [Google Scholar] [CrossRef] [Green Version]
- Sand, L.G.; Scotlandi, K.; Berghuis, D.; Snaar-Jagalska, B.E.; Picci, P.; Schmidt, T.; Szuhai, K.; Hogendoorn, P.C. CXCL14, CXCR7 expression and CXCR4 splice variant ratio associate with survival and metastases in Ewing sarcoma patients. Eur. J. Cancer 2015, 51, 2624–2633. [Google Scholar] [CrossRef] [Green Version]
- Russell-Goldman, E.; Hornick, J.L.; Qian, X.; Jo, V.Y. NKX2.2 immunohistochemistry in the distinction of Ewing sarcoma from cytomorphologic mimics: Diagnostic utility and pitfalls. Cancer Cytopathol. 2018, 126, 942–949. [Google Scholar] [CrossRef] [Green Version]
- Rocchi, A.; Manara, M.C.; Sciandra, M.; Zambelli, D.; Nardi, F.; Nicoletti, G.; Garofalo, C.; Meschini, S.; Astolfi, A.; Colombo, M.P.; et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J. CLIN. Investig. 2010, 120, 668–680. [Google Scholar] [CrossRef]
- van Maldegem, A.M.; Bovee, J.V.; Peterse, E.F.; Hogendoorn, P.C.; Gelderblom, H. Ewing sarcoma: The clinical relevance of the insulin-like growth factor 1 and the poly-ADP-ribose-polymerase pathway. Eur. J. Cancer 2016, 53, 171–180. [Google Scholar] [CrossRef]
- Owen, L.A.; Kowalewski, A.A.; Lessnick, S.L. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS ONE 2008, 3, e1965. [Google Scholar] [CrossRef] [Green Version]
- Infanger, M.; Ulbrich, C.; Baatout, S.; Wehland, M.; Kreutz, R.; Bauer, J.; Grosse, J.; Vadrucci, S.; Cogoli, A.; Derradji, H.; et al. Modeled gravitational unloading induced downregulation of endothelin-1 in human endothelial cells. J. Cell Biochem. 2007, 101, 1439–1455. [Google Scholar] [CrossRef] [PubMed]
- Bartolazzi, A.; Peach, R.; Aruffo, A.; Stamenkovic, I. Interaction between CD44 and hyaluronate is directly implicated in the regulation of tumor development. J. Exp. Med. 1994, 180, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Sy, M.S.; Guo, Y.J.; Stamenkovic, I. Distinct effects of two CD44 isoforms on tumor growth in vivo. J. Exp. Med. 1991, 174, 859–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetler-Stevenson, W.G.; Aznavoorian, S.; Liotta, L.A. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell Biol. 1993, 9, 541–573. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Stamenkovic, I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000, 14, 163–176. [Google Scholar]
- Chetty, C.; Vanamala, S.K.; Gondi, C.S.; Dinh, D.H.; Gujrati, M.; Rao, J.S. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell. Signal. 2012, 24, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Lemma, S.; Avnet, S.; Salerno, M.; Chano, T.; Baldini, N. Identification and validation of housekeeping genes for gene expression analysis of cancer stem cells. PloS ONE 2016, 11, e0149481. [Google Scholar] [CrossRef] [Green Version]
- Elzi, D.J.; Song, M.; Houghton, P.J.; Chen, Y.; Shiio, Y. The role of FLI-1-EWS, a fusion gene reciprocal to EWS-FLI-1, in Ewing sarcoma. Genes Cancer 2015, 6, 452–461. [Google Scholar]
- Ranieri, D.; Proietti, S.; Dinicola, S.; Masiello, M.G.; Rosato, B.; Ricci, G.; Cucina, A.; Catizone, A.; Bizzarri, M.; Torrisi, M.R. Simulated microgravity triggers epithelial mesenchymal transition in human keratinocytes. Sci. Rep. 2017, 7, 538. [Google Scholar] [CrossRef]
- Knowlden, J.M.; Gee, J.M.W.; Seery, L.T.; Farrow, L.; Gullick, W.J.; Ellis, I.O.; Blamey, R.W.; Robertson, J.F.R.; Nicholson, R.I. c- erb B3 and c- erb B4 expression is a feature of the endocrine responsive phenotype in clinical breast cancer. Oncogene 1998, 17, 1949. [Google Scholar] [CrossRef] [Green Version]
- Kreppel, M.; Aryee, D.N.T.; Schaefer, K.-L.; Amann, G.; Kofler, R.; Poremba, C.; Kovar, H. Suppression of KCMF1 by constitutive high CD99 expression is involved in the migratory ability of Ewing’s sarcoma cells. Oncogene 2006, 25, 2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Conza, G.; Buttarelli, M.; Monti, O.; Pellegrino, M.; Mancini, F.; Pontecorvi, A.; Scotlandi, K.; Moretti, F. IGF-1R/MDM2 relationship confers enhanced sensitivity to RITA in Ewing sarcoma cells. Mol. Cancer Ther. 2012, 11, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Nossol, C.; Diesing, A.-K.; Walk, N.; Faber-Zuschratter, H.; Hartig, R.; Post, A.; Kluess, J.; Rothkötter, H.-J.; Kahlert, S. Air–liquid interface cultures enhance the oxygen supply and trigger the structural and functional differentiation of intestinal porcine epithelial cells (IPEC). Histochem. Cell Biol. 2011, 136, 103–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.-Q.; Zhang, S.; Li, S.; An, M.-X.; Li, C.; Yan, J.; Wang, J.-M.; Wang, H.-Q. BAG3 promotes stem cell-like phenotype in breast cancer by upregulation of CXCR4 via interaction with its transcript. Cell Death Dis. 2017, 8, e2933. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Gene | Control (Set as 1) | Spheroid Under s-µg | Adherent Under s-µg | ||
|---|---|---|---|---|---|
| EWS/FLI1 | - | 18.5x * | ↑ | 8.2x * | ↑ |
| CXCR4 | - | 27.2x * | ↑ | 0.9x | = |
| CD 44 | - | 1.5x | ↑ | 3.7x * | ↑ |
| CAV 1 | - | 1.2x * | ↑ | 1.3x * | ↑ |
| VEGF-A | - | 0.9x | = | 0.7x * | ↓ |
| DKK2 | - | 0.8x * | ↓ | 0.9x | = |
| LOX | - | 0.6x | ↓ | 0.6x | ↓ |
| MMP9 | - | 0.9x | = | 1.0x | = |
| ERBB4 | - | 0.8x | ↓ | 0.9x | = |
| NKX2.2 | - | 1.2x | ↑ | 0.9x | = |
| CD99 | - | 1.0x | = | 1.0x | = |
| ZEB2 | - | 1.0x | = | 1.0x | = |
| CXCR7 | - | 1.0x | = | 1.1x | = |
| IGFR1 | - | 0.8x | ↓ | 0.8x | ↓ |
| Gene | Sequences |
|---|---|
| EWS/FLI1 [119] | fwd 5’ GCACCTCCATCCTACCCTCCT 3’ rev 5’ TGGCAGTGGGTGGGTCTTCAT 3’ |
| LOX | fwd 5’ GGCGACGACCCTTACAACC 3’ rev 5’ CTGGGAGACCGTACTGGAAGT 3’ |
| CD44 [24] | fwd 5’ ACCCTCCCCTCATTCACCAT 3’ rev 5’ GTTGTACTACTAGGAGTTGCCTGGATT 3’ |
| MMP9 [120] | fwd 5’ CGCGCTGGGCTTAGATCATT 3’ rev 5’ GGGCGAGGACCATAGAGGT 3’ |
| ZEB2 [120] | fwd 5’ AAGCCAGGGACAGATCAGC 3’ rev 5’ CCACACTCTGTGCATTTGAACT 3’ |
| NKX.2.2 | fwd 5’ GCCCGAGCCAGCCAAGAGG 3’ rev 5’ GCCAGACCGTGCAGGGAGTA 3’ |
| ERBB 4 [121] | fwd 5’ TGTGAGAAGATGGAAGATGGC 3’ rev 5’ GTTGTGGTAAAGTGGAATGGC 3’ |
| CAV 1 [24] | fwd 5’ GTACGACGCGCACACCAA 3’ rev 5’ TCCCTTCTGGTTCTGCAATCA 3’ |
| VEGF-A [24] | fwd 5’ GCGCTGATAGACATCCATGAAC 3’ rev 5’ CTACCTCCACCATGCCAAGTG 3’ |
| E-Cadherin | fwd 5’ CCCGGGACAACGTTTATTAC 3’ rev 5’ GCTGGCTCAAGTCAAAGTCC 3’ |
| CD99 [122] | fwd 5’ TAGGAGATGCTGTTGTTGATGGA 3’ rev 5’ GGATTTGGCATCGGTTTGG 3’ |
| CXCR 4 | fwd 5’ CACTCCCGCCCAATATACCC 3’ rev 5’ TCTGAAGTGTATATCATTCTGGGCT 3’ |
| CXCR 7 | fwd 5’ CAGCAGAGCTCACAGTTGTTG 3’ rev 5’ CGGGCAATCAAATGACCTCC 3’ |
| IGF1-R [123] | fwd 5’ CCATTCTCATGCCTTGGTCT 3’ rev 5’ TGCAAGTTCTGGTTGTCGAG 3’ |
| HPRT1 (Housekeeper) [118] | fwd 5’ TGACCTTGATTTATTTTGCATACC 3’ rev 5’ CGAGCAAGACGTTCAGTCCT 3’ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romswinkel, A.; Infanger, M.; Dietz, C.; Strube, F.; Kraus, A. The Role of C-X-C Chemokine Receptor Type 4 (CXCR4) in Cell Adherence and Spheroid Formation of Human Ewing’s Sarcoma Cells under Simulated Microgravity. Int. J. Mol. Sci. 2019, 20, 6073. https://doi.org/10.3390/ijms20236073
Romswinkel A, Infanger M, Dietz C, Strube F, Kraus A. The Role of C-X-C Chemokine Receptor Type 4 (CXCR4) in Cell Adherence and Spheroid Formation of Human Ewing’s Sarcoma Cells under Simulated Microgravity. International Journal of Molecular Sciences. 2019; 20(23):6073. https://doi.org/10.3390/ijms20236073
Chicago/Turabian StyleRomswinkel, Alexander, Manfred Infanger, Carlo Dietz, Florian Strube, and Armin Kraus. 2019. "The Role of C-X-C Chemokine Receptor Type 4 (CXCR4) in Cell Adherence and Spheroid Formation of Human Ewing’s Sarcoma Cells under Simulated Microgravity" International Journal of Molecular Sciences 20, no. 23: 6073. https://doi.org/10.3390/ijms20236073
APA StyleRomswinkel, A., Infanger, M., Dietz, C., Strube, F., & Kraus, A. (2019). The Role of C-X-C Chemokine Receptor Type 4 (CXCR4) in Cell Adherence and Spheroid Formation of Human Ewing’s Sarcoma Cells under Simulated Microgravity. International Journal of Molecular Sciences, 20(23), 6073. https://doi.org/10.3390/ijms20236073