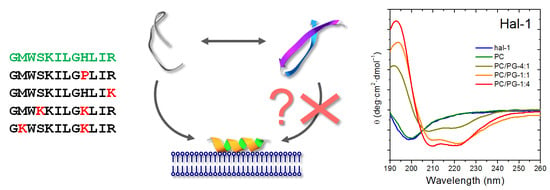
Interaction of Halictine-Related Antimicrobial Peptides with Membrane Models
Abstract
:1. Introduction
2. Results and Discussion
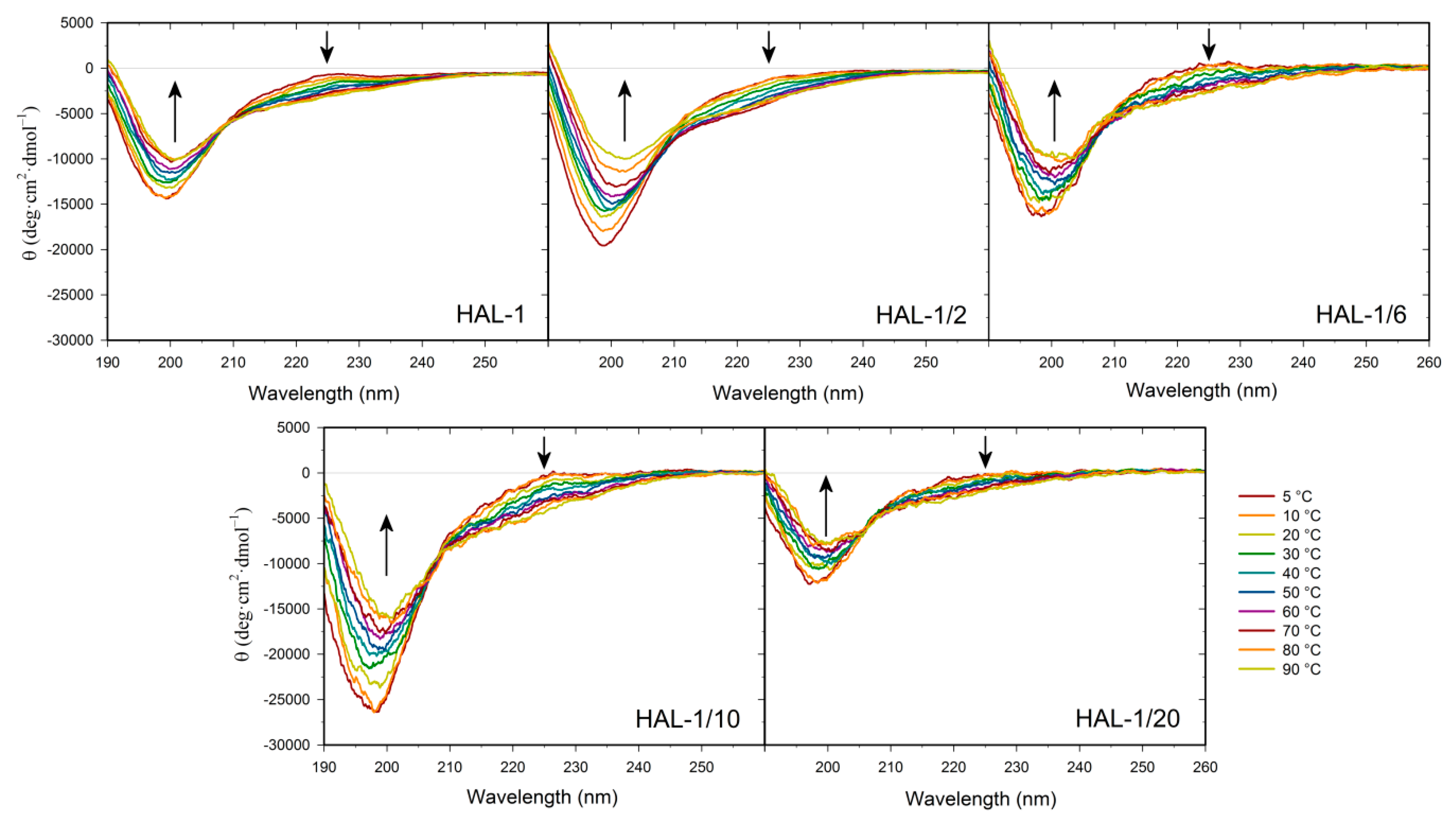
2.1. Structural Changes Followed by ECD
2.1.1. Structural Changes Due to the Presence of TFE and SDS
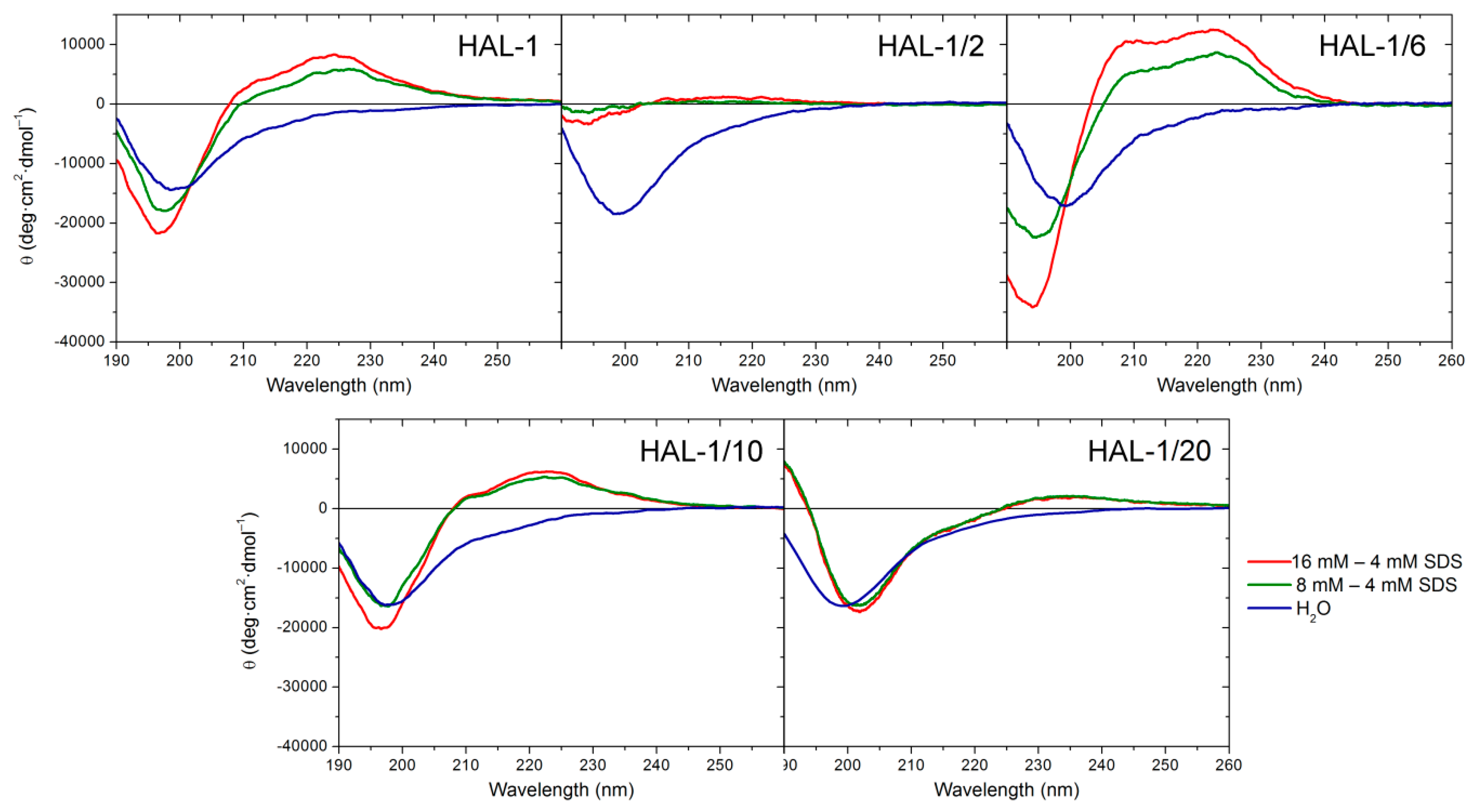
2.1.2. Structural Changes Due to the Presence of LUVs
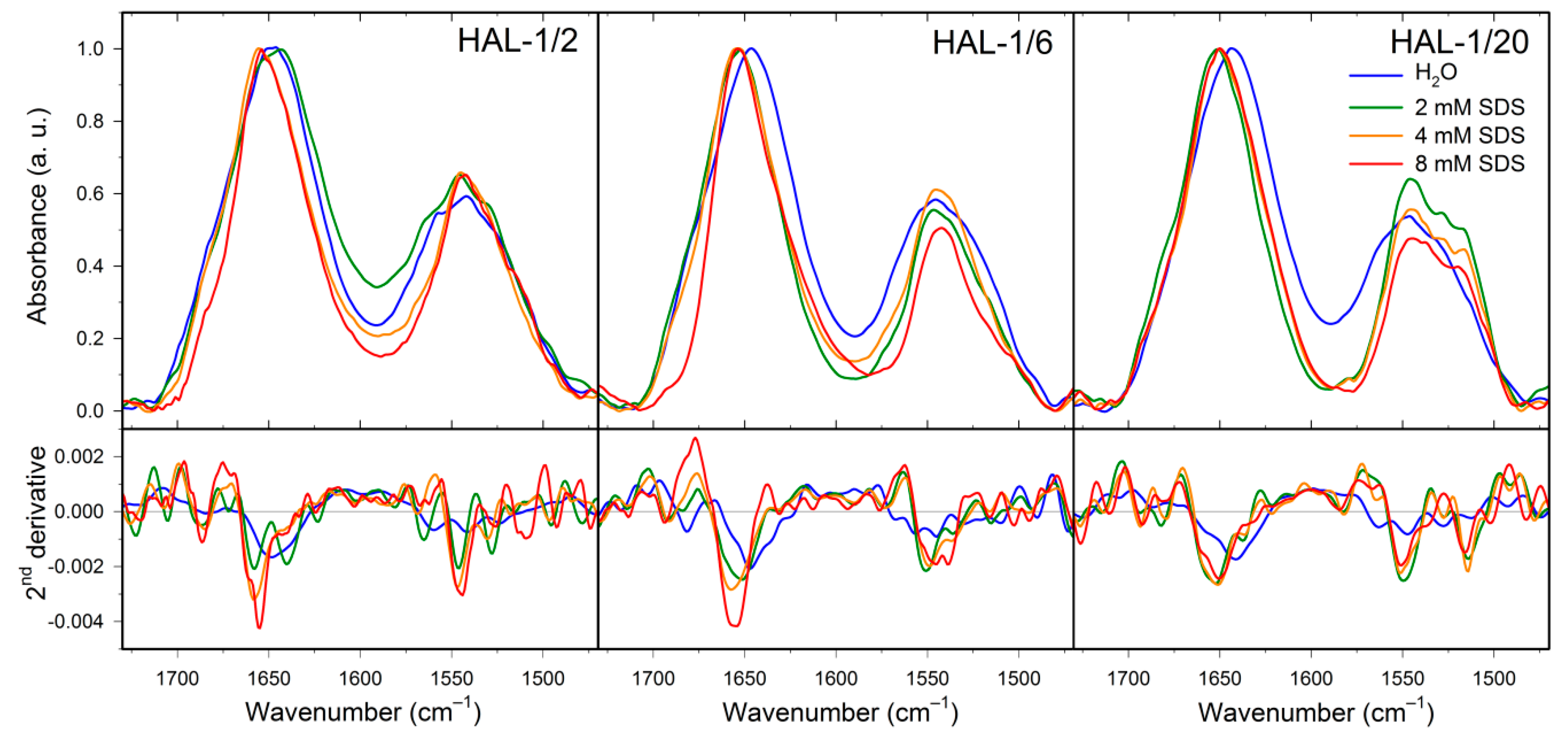
2.2. Structural Changes Followed by Infrared Spectroscopy
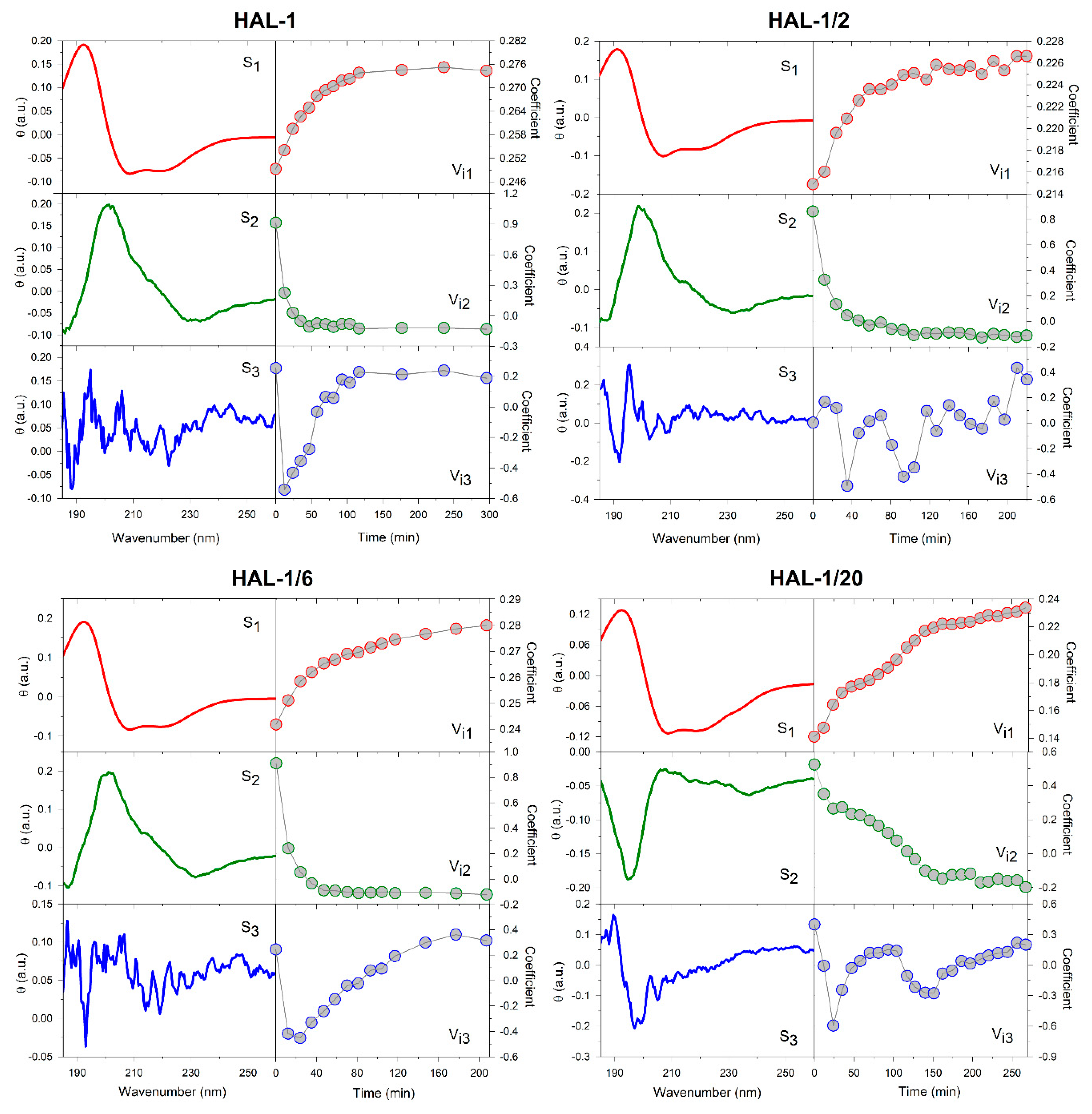
2.3. Concentration Dependence Measurements
2.4. Structural Changes Followed by Fluorescence Spectroscopy
2.5. Molecular Dynamics
3. Materials and Methods
3.1. Materials
3.2. Preparation of Vesicles
3.3. Electronic Circular Dichroism
3.4. Principle Component Analysis
3.5. Infrared Spectroscopy
3.6. Vibrational Circular Dichroism
3.7. Fluorescence Spectroscopy
3.8. Molecular Dynamics
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMPs | Antimicrobial peptides |
| cmc | Critical micelle concentration |
| ECD | Electronic circular dichroism |
| HAL | Halictine |
| IR | Infrared |
| L/P | Lipid/peptide ratio |
| LUV | Large unilamellar vesicle |
| MD | Molecular dynamics |
| PC | 1,2-Dimyristoyl-sn-glycerol-3-phosphatidylcholine |
| PCA | Principal component analysis |
| PG | 1,2-Dimyristoyl-sn-glycero-3-phospho-(1′-rac-glycerol) |
| PPII | Polyproline II |
| SDS | Sodium dodecyl sulfate |
| TFE | 2,2,2-Trifluoroethanol |
| VCD | Vibrational circular dichroism |
Appendix A
| Solution | Hal-1 | Hal-1/2 | Hal-1/6 | Hal-1/10 | Hal-1/20 |
|---|---|---|---|---|---|
| Water | 14% | 14% | 13% | 15% | 15% |
| TFE 30% | 32% | 32% | 37% | 36% | 36% |
| TFE 50% | 32% | 32% | 40% | 42% | 36% |
| SDS 0.016 mM | 13% | 13% | 3% | 14% | 14% |
| SDS 0.16 mM | 14% | 14% | 3% | 18% | 52% |
| SDS 2 mM | 46% | 46% | 66% | 63% | 36% |
| SDS 4 mM | 51% | 51% | 46% | 63% | 32% |
| SDS 8 mM | 37% | 37% | 46% | 50% | 31% |
| SDS 16 mM | 31% | 31% | 36% | 36% | 42% |
| Structure | Sodium Dodecyl Sulfate | ||||||
|---|---|---|---|---|---|---|---|
| 0 mM | 0.016 mM | 0.16 mM | 2 mM | 4 mM | 8 mM | 16 mM | |
| HAL-1 | |||||||
| α-helix | 12% | 12% | 14% | 61% | 61% | 53% | 53% |
| β-sheet | 50% | 50% | 44% | 6% | 6% | 10% | 10% |
| β-turn | 16% | 16% | 16% | 12% | 12% | 17% | 17% |
| Other | 22% | 22% | 26% | 22% | 22% | 21% | 21% |
| HAL-1/2 | |||||||
| α-helix | 12% | 11% | 14% | 14% | 47% | 47% | 44% |
| β-sheet | 53% | 54% | 45% | 44% | 13% | 13% | 15% |
| β-turn | 16% | 16% | 16% | 16% | 18% | 18% | 18% |
| Other | 19% | 19% | 25% | 25% | 22% | 22% | 24% |
| HAL-1/6 | |||||||
| α-helix | 12% | 13% | 11% | 82% | 85% | 70% | 60% |
| β-sheet | 52% | 42% | 37% | 2% | 2% | 4% | 9% |
| β-turn | 16% | 16% | 14% | 9% | 9% | 14% | 19% |
| Other | 20% | 29% | 38% | 6% | 4% | 11% | 14% |
| HAL-1/10 | |||||||
| α-helix | 11% | 12% | 13% | 80% | 79% | 72% | 70% |
| β-sheet | 52% | 51% | 43% | 2% | 2% | 4% | 5% |
| β-turn | 16% | 16% | 16% | 10% | 10% | 14% | 15% |
| Other | 21% | 21% | 27% | 8% | 9% | 10% | 10% |
| HAL-1/20 | |||||||
| α-helix | 12% | 13% | 66% | 42% | 39% | 37% | 60% |
| β-sheet | 52% | 42% | 5% | 13% | 17% | 18% | 18% |
| β-turn | 16% | 13% | 12% | 17% | 18% | 18% | 17% |
| Other | 20% | 32% | 18% | 28% | 27% | 27% | 15% |
| Structure | LUV | |||||
|---|---|---|---|---|---|---|
| 0 mM | PC (L/P = 20) | PC (L/P = 100) | PC/PG (4:1) (L/P = 20) | PC/PG (1:1) (L/P = 20) | PC/PG (1:4) (L/P = 20) | |
| HAL-1 | ||||||
| α-helix | 12% | 12% | 18% | 35% | 45% | 61% |
| β-sheet | 50% | 51% | 38% | 17% | 10% | 7% |
| β-turn | 16% | 16% | 18% | 18% | 17% | 12% |
| Other | 22% | 21% | 26% | 30% | 22% | 21% |
| HAL-1/2 | ||||||
| α-helix | 11% | 11% | 17% | 37% | 47% | 69% |
| β-sheet | 53% | 53% | 40% | 17% | 10% | 5% |
| β-turn | 16% | 16% | 18% | 17% | 16% | 14% |
| Other | 19% | 21% | 25% | 29% | 28% | 13% |
| HAL-1/6 | ||||||
| α-helix | 12% | 12% | 14% | 60% | 79% | 60% |
| β-sheet | 52% | 50% | 45% | 7% | 1% | 6% |
| β-turn | 16% | 16% | 17% | 8% | 5% | 12% |
| Other | 20% | 22% | 24% | 25% | 14% | 21% |
| HAL-1/10 | ||||||
| α-helix | 11% | 12% | 13% | 35% | 69% | 36% |
| β-sheet | 52% | 52% | 51% | 18% | 3% | 38% |
| β-turn | 16% | 16% | 17% | 17% | 9% | 14% |
| Other | 21% | 20% | 19% | 30% | 7% | 13% |
| HAL-1/20 | ||||||
| α-helix | 12% | 12% | 14% | 25% | 80% | 46% |
| β-sheet | 52% | 50% | 46% | 24% | 2% | 13% |
| β-turn | 16% | 16% | 17% | 18% | 10% | 18% |
| Other | 20% | 21% | 22% | 34% | 7% | 24% |
| Solvent | Amide I (cm−1) | Second Derivative Decomposition of Amide I (cm−1) | ||||||
|---|---|---|---|---|---|---|---|---|
| aggregate | β-sheet | α-helix | 310-helix | coil | β-turn | β-sheet | ||
| HAL-1 | ||||||||
| H2O | 1646 | — | — | — | — | 1648 (88) | 1683 (12) | — |
| D2O | 1647 | — | — | — | — | 1642(69) | 1667 (31) | — |
| TFE | 1655 | — | 1633 (26) | 1656 (63) | — | — | 1680 (11) | — |
| SDS 8 mM/H2O | 1655 | 1621 (12) | 1635 (16) | 1656 (63) | — | — | 1682 (9) | — |
| SDS 8 mM/D2O | 1649 | — | — | 1649 (95) | — | — | 1676 (5) | — |
| PC | 1656 | 1619 (5) | — | 1656 (63) | — | — | 1688 (33) | — |
| PC/PG 1:1 | 1655 | — | 1634 (29) | 1656 (52) | — | — | 1680 (19) | — |
| PC/PG 1:4 | 1656 | — | 1633 (6) | 1655 (58) | — | — | 1678 (36) | — |
| HAL-1/2 | ||||||||
| H2O | 1649 | — | 1637 (29) | — | — | 1650 (56) | 1684 (14) | — |
| SDS 8 mM/H2O | 1655 | — | 1635(38) | 1657 (58) | — | — | 1685 (4) | — |
| PC | 1657 | — | 1642 (32) | 1657 (32) | — | — | 1680 (35) | — |
| PC/PG 1:1 | 1647 | 1628 (34) | 1642 (28) | 1658 (24) | — | — | 1676 (13) | — |
| PC/PG 1:4 | 1657 | — | 1632 (15) | 1656 (43) | — | — | 1680 (41) | — |
| HAL-1/6 | ||||||||
| H2O | 1646 | — | — | — | — | 1647 (92) | 1682 (8) | — |
| SDS 8 mM/H2O | 1654 | 1629 (29) | — | 1655 (69) | — | — | — | 1695 (2) |
| PC | 1654 | 1621 (7) | 1639 (29) | 1657 (59) | — | — | 1680 (5) | — |
| PC/PG 1:1 | 1656 | 1626 (9) | 1642 (18) | 1657 (62) | — | — | 1682 (11) | — |
| HAL-1/10 | ||||||||
| H2O | 1647 | — | — | — | — | 1647 (82) | 1681 (18) | — |
| PC/PG 1:1 | 1655 | 1625 (12) | 1640 (21) | 1657 (57) | — | — | 1681 (10) | — |
| PC/PG 1:4 | 1656 | — | — | 1656 (80) | — | — | 1688 (20) | — |
| HAL-1/20 | ||||||||
| H2O | 1644 | — | 1643 (93) | — | — | — | 1676 (7) | — |
| SDS 8 mM/H2O | 1650 | 1629 (9) | — | 1653 (83) | — | — | 1685 (9) | — |
| PC | 1649 | 1625 (36) | 1639 (13) | 1654 (28) | — | — | 1685 (22) | — |
| PC/PG 1:1 | 1650 | 1628 (29) | 1642 (16) | 1659 (49) | — | — | 1685 (6) | — |
| PC/PG 1:4 | 1654 | 1621 (19) | 1634 (14) | 1655 (43) | — | — | 1678 (24) | — |

References
- Phoenix, D.A.; Dennison, S.R.; Harris, F. Antibacterial Peptides; Wiley-VCH: Weinheim, Germany, 2013; ISBN 978-3-527-33263-2. [Google Scholar]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.; Schneider, G. Designing antimicrobial peptides: Form follows function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Hancock, R.E.; Scott, M.G. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA 2000, 97, 8856–8861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. 2005, 62, 784–790. [Google Scholar] [CrossRef]
- Papo, N.; Shai, Y. Can we predict biological activity of antimicrobial peptides from their interactions with model phospholipid membranes? Peptides 2003, 24, 1693–1703. [Google Scholar] [CrossRef]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef]
- Yount, N.Y.; Yeaman, M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2004, 101, 7363–7368. [Google Scholar] [CrossRef]
- Epand, R.M.; Epand, R.F. Domains in bacterial membranes and the action of antimicrobial agents. Mol. Biosyst. 2009, 5, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Epand, R.F.; Arnusch, C.J.; Papahadjopoulos-Sternberg, B.; Wang, G.; Shai, Y. Lipid clustering by three homologous arginine-rich antimicrobial peptides is insensitive to amino acid arrangement and induced secondary structure. Biochim. Biophys. Acta 2010, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C.; Hristova, K. Antimicrobial peptides: Successes, challenges and unanswered questions. J. Membr. Biol. 2011, 239, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Blondelle, S.E.; Lohner, K.; Aguilar, M.I. Lipid-induced conformation and lipid-binding properties of cytolytic and antimicrobial peptides: Determination and biological specificity. Biochim. Biophys. Acta 1999, 1462, 89–108. [Google Scholar] [CrossRef]
- Epand, R.F.; Mor, A.; Epand, R.M. Lipid complexes with cationic peptides and OAKs; their role in antimicrobial action and in the delivery of antimicrobial agents. Cell. Mol. Life Sci. 2011, 68, 2177–2188. [Google Scholar] [CrossRef]
- Čeřovský, V.; Buděšínský, M.; Hovorka, O.; Cvačka, J.; Voburka, Z.; Slaninová, J.; Borovičková, L.; Fučík, V.; Bednárová, L.; Votruba, I.; et al. Lasioglossins: Three novel antimicrobial peptides from the venom of the eusocial bee Lasioglossum laticeps (Hymenoptera: Halictidae). ChemBioChem 2009, 10, 2089–2099. [Google Scholar] [CrossRef]
- Čeřovský, V.; Slaninová, J.; Fučík, V.; Monincová, L.; Bednárová, L.; Maloň, P.; Stokrová, J. Lucifensin, a novel insect defensin of medicinal maggots: Synthesis and structural study. ChemBioChem 2011, 12, 1352–1361. [Google Scholar] [CrossRef]
- Monincová, L.; Buděšínský, M.; Slaninová, J.; Hovorka, O.; Cvačka, J.; Voburka, Z.; Fučík, V.; Borovičková, L.; Bednárová, L.; Straka, J.; et al. Novel antimicrobial peptides from the venom of the eusocial bee Halictus sexcinctus (Hymenoptera: Halictidae) and their analogs. Amino Acids 2010, 39, 763–775. [Google Scholar] [CrossRef]
- Čujová, S.; Bednárová, L.; Slaninová, J.; Straka, J.; Čeřovský, V. Interaction of a novel antimicrobial peptide isolated from the venom of solitary bee Colletes daviesanus with phospholipid vesicles and Escherichia coli cells. J. Pept. Sci. 2014, 20, 885–895. [Google Scholar] [CrossRef]
- Stanchev, S.; Zawada, Z.; Monincová, L.; Bednárová, L.; Slaninová, J.; Fučík, V.; Čeřovský, V. Synthesis of lucifensin by native chemical ligation and characteristics of its isomer having different disulfide bridge pattern. J. Pept. Sci. 2014, 20, 725–735. [Google Scholar] [CrossRef]
- Nešuta, O.; Hexnerová, R.; Buděšínský, M.; Slaninová, J.; Bednárová, L.; Hadravová, R.; Straka, J.; Veverka, V.; Čeřovský, V. Antimicrobial peptide from the wild bee Hylaeus signatus venom and its analogues: Structure-activity study and synergistic effect with antibiotics. J. Nat. Prod. 2016, 79, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Pazderková, M.; Kočišová, E.; Pazderka, T.; Maloň, P.; Kopecký, V.; Monincová, L.; Čeřovský, V.; Bednárová, L. Antimicrobial peptide from the eusocial bee Halictus sexcinctus interacting with model membranes. In Advances in Biomedical Spectroscopy; Marques, M.P., Batista de Carvalho, L.A.E., Haris, P.I., Eds.; IOE Press: Amsterdam, The Netherlands, 2013; Volume 7, pp. 79–83. ISBN 978-1-61499-183-0. [Google Scholar]
- Myers, J.K.; Pace, C.N.; Scholtz, J.M. Trifluoroethanol effects on helix propensity and electrostatic interactions in the helical peptide from ribonuclease T-1. Protein Sci. 1998, 7, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Warschawski, D.E.; Arnold, A.A.; Beaugrand, M.; Gravel, A.; Chartrand, É.; Marcotte, I. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. Biochim. Biophys. Acta 2011, 1808, 1957–1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strandberg, E.; Tiltak, D.; Ehni, S.; Wadhwani, P.; Ulrich, A.S. Lipid shape is a key factor for membrane interactions of amphipathic helical peptides. Biochim. Biophys. Acta 2012, 1818, 1764–1776. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, D.; Weiss, R.M.; Terwilliger, T.C. The helical hydrophobic moment: A measure of the amphiphilicity of a helix. Nature 1982, 277, 371–374. [Google Scholar] [CrossRef]
- Thévenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tufféry, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Backlund, B.; Wikander, G.; Peeters, T.L.; Gräslund, A. Induction of secondary structure in the peptide hormone motilin by interaction with phospholipid vesicles. Biochim. Biophys. Acta 1994, 1190, 337–344. [Google Scholar] [CrossRef]
- Drake, A.F.; Siligardi, G.; Gibbons, W.A. Reassessment of the electronic circular dichroism criteria for random coil conformations of poly(L-lysine) and the implications for protein folding and denaturation studies. Biophys. Chem. 1988, 31, 143–146. [Google Scholar] [CrossRef]
- Wimmer, R.; Andersen, K.K.; Vad, B.; Davidsen, M.; Mølgaard, S.; Nesgaard, L.W.; Kristensen, H.H.; Otzen, D.E. Versatile interactions of the antimicrobial peptide Novispirin with detergents and lipids. Biochemistry 2006, 45, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta 2004, 1666, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Berova, N.; Polavarapu, P.L.; Nakanishi, K.; Woody, R.W. Comprehensive Chiroptical Spectroscopy Applications in Stereochemical Analysis of Synthetic Compounds, Natural Products and Biomolecules; John Wiley & Sons: Hoboken, NJ, USA, 2012; ISBN 978-1-118-01292-5. [Google Scholar]
- Abdul-Gader, A.; Miles, A.J.; Wallace, B.A. A reference dataset for the analyses of membrane protein secondary structures and transmembrane residues using circular dichroism spectroscopy. Bioinformatics 2011, 27, 1630–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreerama, N.; Woody, R.W. Poly(Pro)II helices in globular proteins: Identification and circular dichroic analysis. Biochemistry 1994, 33, 10022–10025. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Woollett, B.; Miles, A.J.; Janes, R.W.; Wallace, B.A. The protein circular dichroism data bank, a Web-based site for access to circular dichroism spectroscopic data. Structure 2010, 18, 1267–1269. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.C. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 1999, 35, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Bochicchio, B.; Tamburro, A.M. Polyproline II structure in proteins: Identification by chiroptical spectroscopies, stability, and functions. Chirality 2002, 14, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.L.S.; Miles, A.J.; Whitmore, L.; Wallace, B.A. Distinct circular dichroism spectroscopic signatures of polyproline II and unordered secondary structures: Applications in secondary structure analyses. Protein Sci. 2014, 23, 1765–1772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Tesař, A.; Kopecký Jr., V.; Kočišová, E.; Bednárová, L. Dynamics of lipid layers with/without bounded antimicrobial peptide halictine-1. Vibrat. Spectrosc. 2017, 93, 42–51. [Google Scholar] [CrossRef]
- Miller, L.M.; Bourassa, M.W.; Smith, R.J. FTIR spectroscopic imaging of protein aggregation in living cells. Biochim. Biophys. Acta 2013, 1828, 2339–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, M.; Mantsch, H.H. The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Keiderling, T.A.; Silva, R.A.; Yoder, G.; Dukor, R.K. Vibrational circular dichroism spectroscopy of selected oligopeptide conformations. Bioorg. Med. Chem. 1999, 7, 133–141. [Google Scholar] [CrossRef]
- Keiderling, T.A. Protein and peptide secondary structure and conformational determination with vibrational circular dichroism. Curr. Opin. Chem. Biol. 2002, 6, 682–688. [Google Scholar] [CrossRef]
- Ma, S.; Freedman, T.B.; Dukor, R.K.; Nafie, L.A. Near-infrared and mid-infrared Fourier transform vibrational circular dichroism of proteins in aqueous solution. Appl. Spectrosc. 2010, 64, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Cao, X.; Mak, M.; Sadik, A.; Walkner, C.; Freedman, T.B.; Lednev, I.K.; Dukor, R.K.; Nafie, L.A. Vibrational circular dichroism shows unusual sensitivity to protein fibril formation and development in solution. J. Am. Chem. Soc. 2007, 129, 12364–12365. [Google Scholar] [CrossRef] [PubMed]
- Kurouski, D.; Lombardi, R.A.; Dukor, R.K.; Lednev, I.K.; Nafie, L.A. Direct observation and pH control of reversed supramolecular chirality in insulin fibrils by vibrational circular dichroism. Chem. Commun. 2010, 46, 7154–7156. [Google Scholar] [CrossRef] [PubMed]
- Measey, T.J.; Schweitzer-Stenner, R. Vibrational circular dichroism as a probe of fibrillogenesis: The origin of the anomalous intensity enhancement of amyloid-like fibrils. J. Am. Chem. Soc. 2011, 133, 1066–1076. [Google Scholar] [CrossRef]
- Krishnakumar, S.S.; London, E. Effect of sequence hydrophobicity and bilayer width upon the minimum length required for the formation of transmembrane helices in membranes. J. Mol. Biol. 2007, 374, 671–687. [Google Scholar] [CrossRef]
- Zeth, K. Structure and mechanism of human antimicrobial peptide dermcidin and its antimicrobial potential. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2013; Volume 2, pp. 1333–1342. ISBN 978-84-942134-0-3. [Google Scholar]
- Huang, H.W. Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochim. Biophys. Acta 2006, 1758, 1292–1302. [Google Scholar] [CrossRef] [Green Version]
- Millhauser, G.L. Views of helical peptides: A proposal for the position of 310-helix along the thermodynamic folding pathway. Biochemistry 1995, 34, 3873–3877. [Google Scholar] [CrossRef] [PubMed]
- Leontiadou, H.; Mark, A.E.; Marrink, S.J. Antimicrobial peptides in action. J. Am. Chem. Soc. 2006, 128, 12156–12161. [Google Scholar] [CrossRef] [PubMed]
- Forbrig, E.; Staffa, J.K.; Salewski, J.; Mroginski, M.A.; Hildebrandt, P.; Kozuch, J. Monitoring the orientational changes of Alamethicin during incorporation into bilayer lipid membranes. Langmuir 2018, 34, 2373–2385. [Google Scholar] [CrossRef] [PubMed]
- Reißer, S.; Strandberg, E.; Steinbrecher, T.; Elstner, M.; Ulrich, A.S. Best of two worlds? How MD simulations of amphiphilic helical peptides in membranes can complement data from oriented solid-state NMR. J. Chem. Theory Comput. 2018, 14, 6002–6014. [Google Scholar] [CrossRef] [PubMed]
- Andruschenko, V.; Vogel, H.J.; Prenner, E.J. Optimization of the hydrochloric acid concentration used for trifluoroacetate removal from synthetic peptides. J. Pept. Sci. 2007, 13, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Mukerjee, P.; Mysels, K.J. Critical Micelle Concentration of Aqueous Surfactant Systems; NSRDS-NBS 36; U.S. Department of Commerce: Washington, DC, USA, 1971.
- Rohl, C.; Baldwin, R.L. Deciphering rules of helix stability in peptides. Methods Enzymol. 1998, 296, 1–26. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004, 383, 318–351. [Google Scholar] [CrossRef]
- Malinowski, E.R. Factor Analysis in Chemistry, 3rd ed.; Wiley: Chichester, UK, 2002; ISBN 0-471-13479-1. [Google Scholar]
- Dousseau, F.; Therrien, M.; Pézolet, M. On the spectral subtraction of water from the FT-IR spectra of aqueous solutions of proteins. Appl. Spectrosc. 1989, 43, 538–542. [Google Scholar] [CrossRef]
- Roux, S.; Zékri, E.; Rousseau, B.; Paternostre, M.; Cintrat, J.C.; Fay, N. Elimination and exchange of trifluoroacetate counter-ion from cationic peptides: A critical evaluation of different approaches. J. Pept. Sci. 2008, 14, 354–359. [Google Scholar] [CrossRef]
- Nafie, L.A.; Buijs, H.; Rilling, A.; Cao, X.; Dukor, R.K. Dual source Fourier transform polarization modulation spectroscopy: An improved method for the measurement of circular and linear dichroism. Appl. Spectrosc. 2004, 58, 647–654. [Google Scholar] [CrossRef]
- Nafie, L.A. Dual polarization modulation: Real-time, spectral multiplex separation of circular dichroism from linear birefringence spectral intensities. Appl. Spectrosc. 2000, 54, 1634–1645. [Google Scholar] [CrossRef]
- Case, D.A.; Darden, T.A.; Cheatham, T.E., III; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 11; University of California: San Francisco, CA, USA, 2010. [Google Scholar]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, S.; Kollman, P.A.; Settle, K. An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 2004, 13, 100–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]








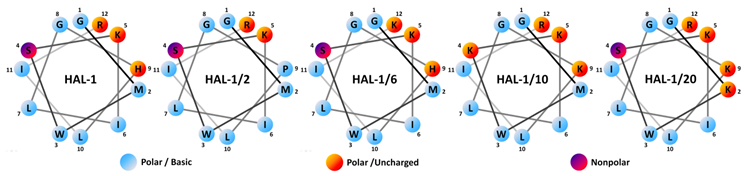
| Acronym | Sequence | MW (Da) | Charge | µH | H | Antimicrobial Activity MIC (µM) | Hemolytic LC50 (µM) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| B.1 | S.2 | E.3 | P.4 | |||||||
| HAL-1 | GMWSKILGHLIR | 1408.9 | +3 | 0.380 | −0.004 | 0.8 | 7.7 | 3.8 | 45.0 | 82 |
| HAL-1/2 | GMWSKILGPLIR | 1368.8 | +3 | 0.361 | +0.023 | 3.6 | >100 | 30.0 | >100 | >200 |
| HAL-1/6 | GMWSKILGHLIK | 1380.6 | +3 | 0.323 | +0.051 | 1.3 | 15.8 | 7.2 | 65.0 | 132 |
| HAL-1/10 | GMWKKILGKLIR | 1440.9 | +5 | 0.416 | –0.133 | 0.8 | 15.0 | 2.3 | 13.1 | >200 |
| HAL-1/20 | GKWSKILGKLIR | 1396.9 | +5 | 0.473 | –0.176 | 1.7 | 21.7 | 2.3 | 28.3. | >200 |
 | ||||||||||
| Solution | HAL-1 | HAL-1/2 | HAL-1/6 | HAL-1/20 |
|---|---|---|---|---|
| Water | 360 nm | 356 nm | 361 nm | 359 nm |
| PC | 356 nm | 362 nm | 356 nm | 359 nm |
| PC/PG (1:1) | 331 nm | 350 nm | 336 nm | 333 nm |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pazderková, M.; Maloň, P.; Zíma, V.; Hofbauerová, K.; Kopecký, V., Jr.; Kočišová, E.; Pazderka, T.; Čeřovský, V.; Bednárová, L. Interaction of Halictine-Related Antimicrobial Peptides with Membrane Models. Int. J. Mol. Sci. 2019, 20, 631. https://doi.org/10.3390/ijms20030631
Pazderková M, Maloň P, Zíma V, Hofbauerová K, Kopecký V Jr., Kočišová E, Pazderka T, Čeřovský V, Bednárová L. Interaction of Halictine-Related Antimicrobial Peptides with Membrane Models. International Journal of Molecular Sciences. 2019; 20(3):631. https://doi.org/10.3390/ijms20030631
Chicago/Turabian StylePazderková, Markéta, Petr Maloň, Vlastimil Zíma, Kateřina Hofbauerová, Vladimír Kopecký, Jr., Eva Kočišová, Tomáš Pazderka, Václav Čeřovský, and Lucie Bednárová. 2019. "Interaction of Halictine-Related Antimicrobial Peptides with Membrane Models" International Journal of Molecular Sciences 20, no. 3: 631. https://doi.org/10.3390/ijms20030631
APA StylePazderková, M., Maloň, P., Zíma, V., Hofbauerová, K., Kopecký, V., Jr., Kočišová, E., Pazderka, T., Čeřovský, V., & Bednárová, L. (2019). Interaction of Halictine-Related Antimicrobial Peptides with Membrane Models. International Journal of Molecular Sciences, 20(3), 631. https://doi.org/10.3390/ijms20030631