Potential for Drug Repositioning of Midazolam for Dentin Regeneration
Abstract
:1. Introduction
2. Results
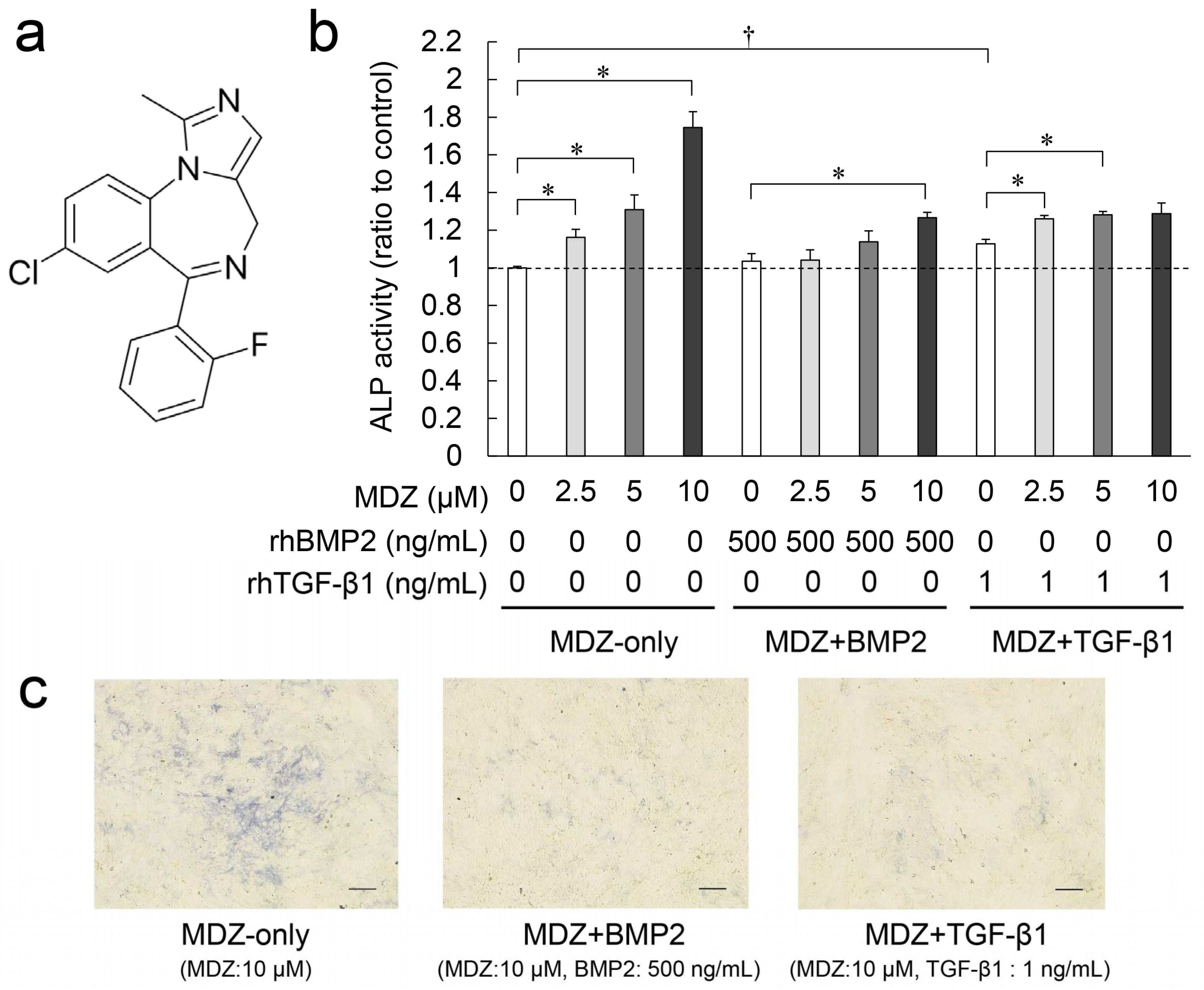
2.1. Combined Effect of MDZ with BMP2 or TGF-β1 on Differentiation of the PPU-7 Cell Line
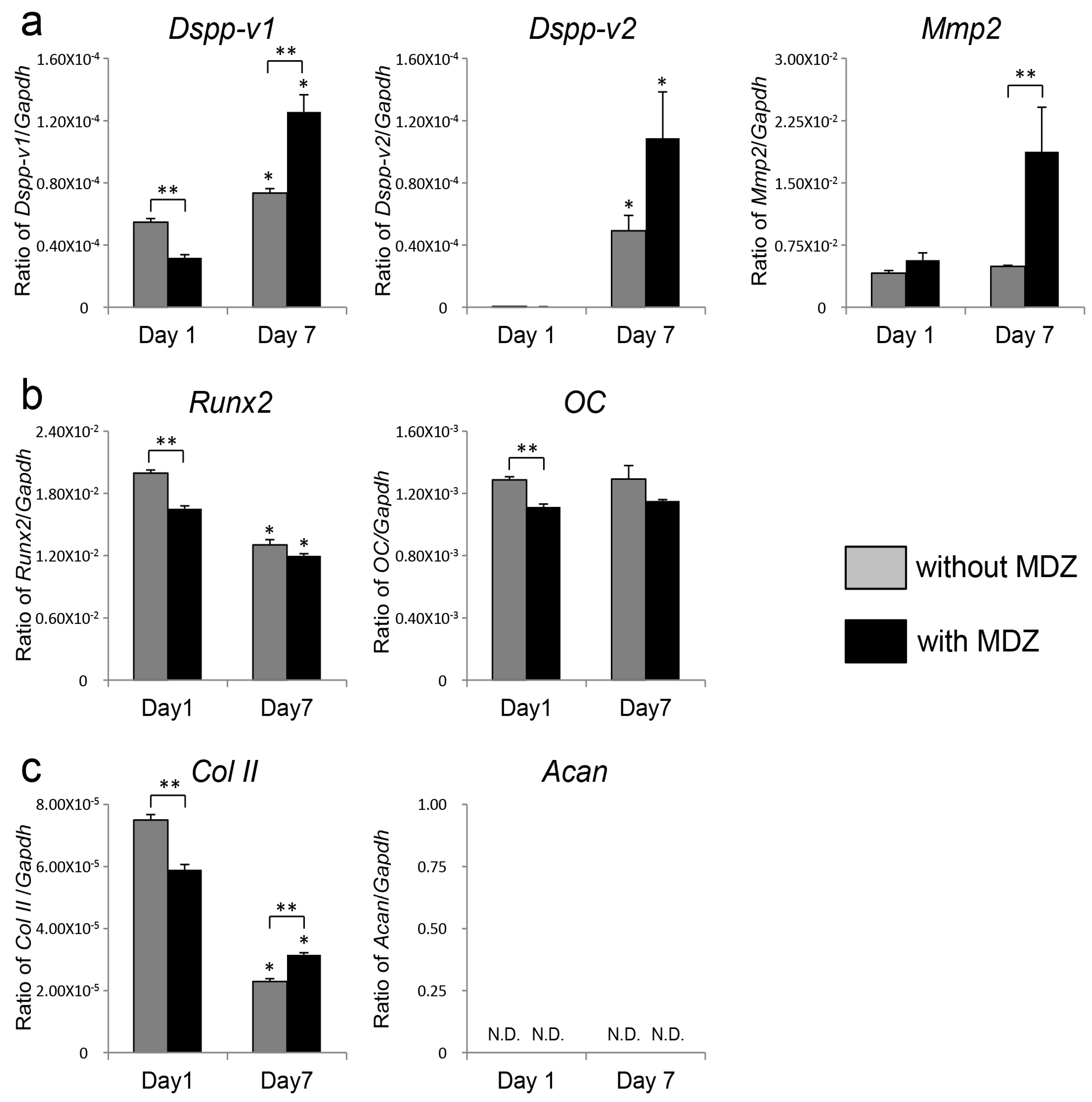
2.2. Effect of MDZ on Temporal Changes in Gene Expression of PPU-7 Cell Line
2.3. Effect of MDZ on Mineralization Induction of the PPU-7 Cell Line
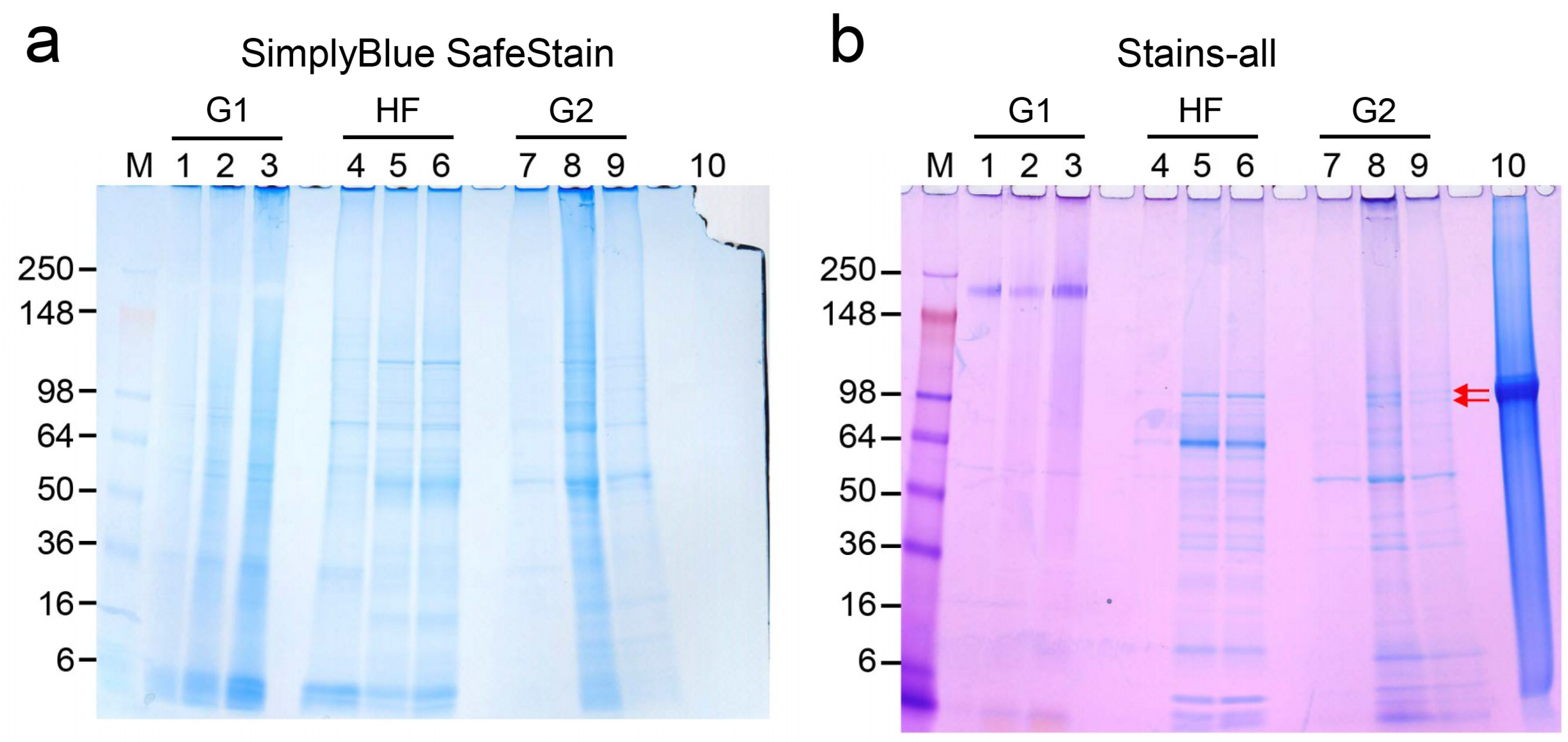
2.4. Detection of DPP in Precipitated Nodules from PPU-7 Cells
2.5. X-ray Diffraction (XRD) Patterns of Precipitated Nodules
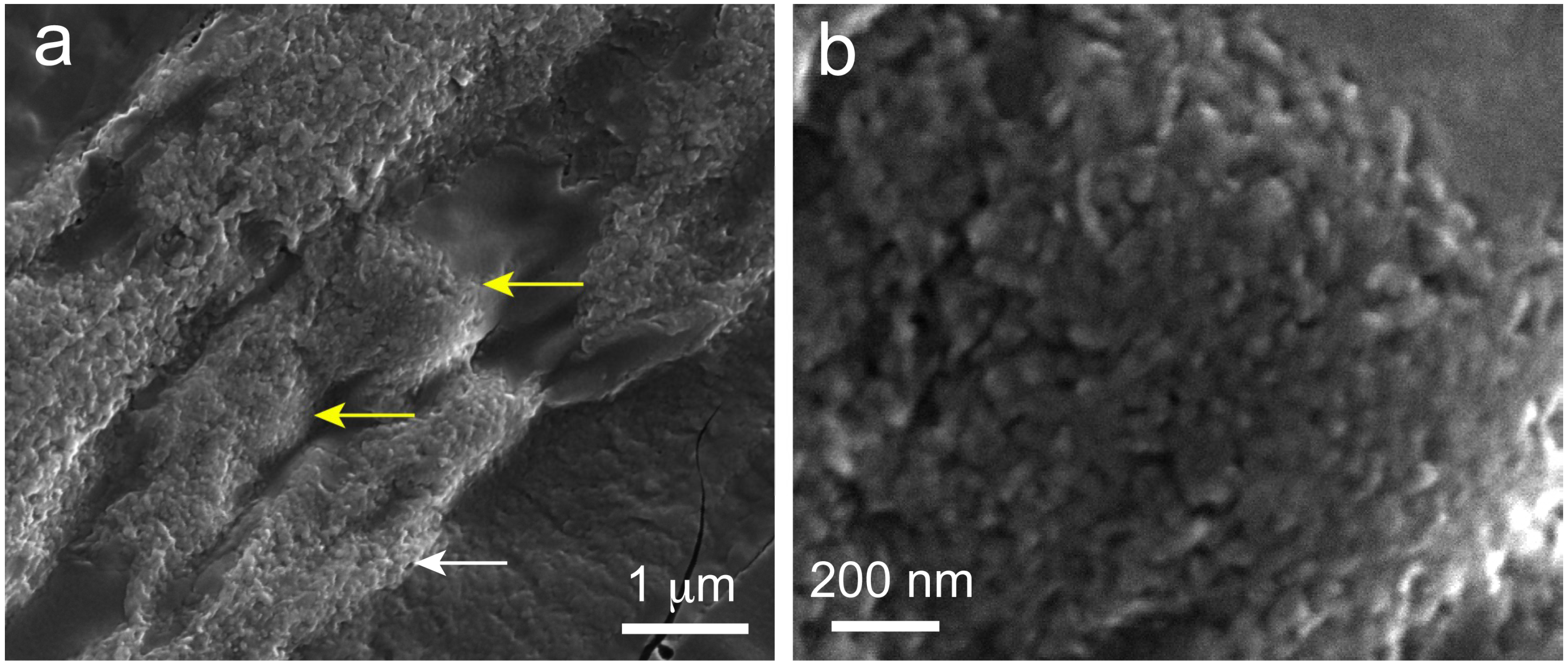
2.6. Scanning Electron Microscopy (SEM) Observations of Precipitated Nodules
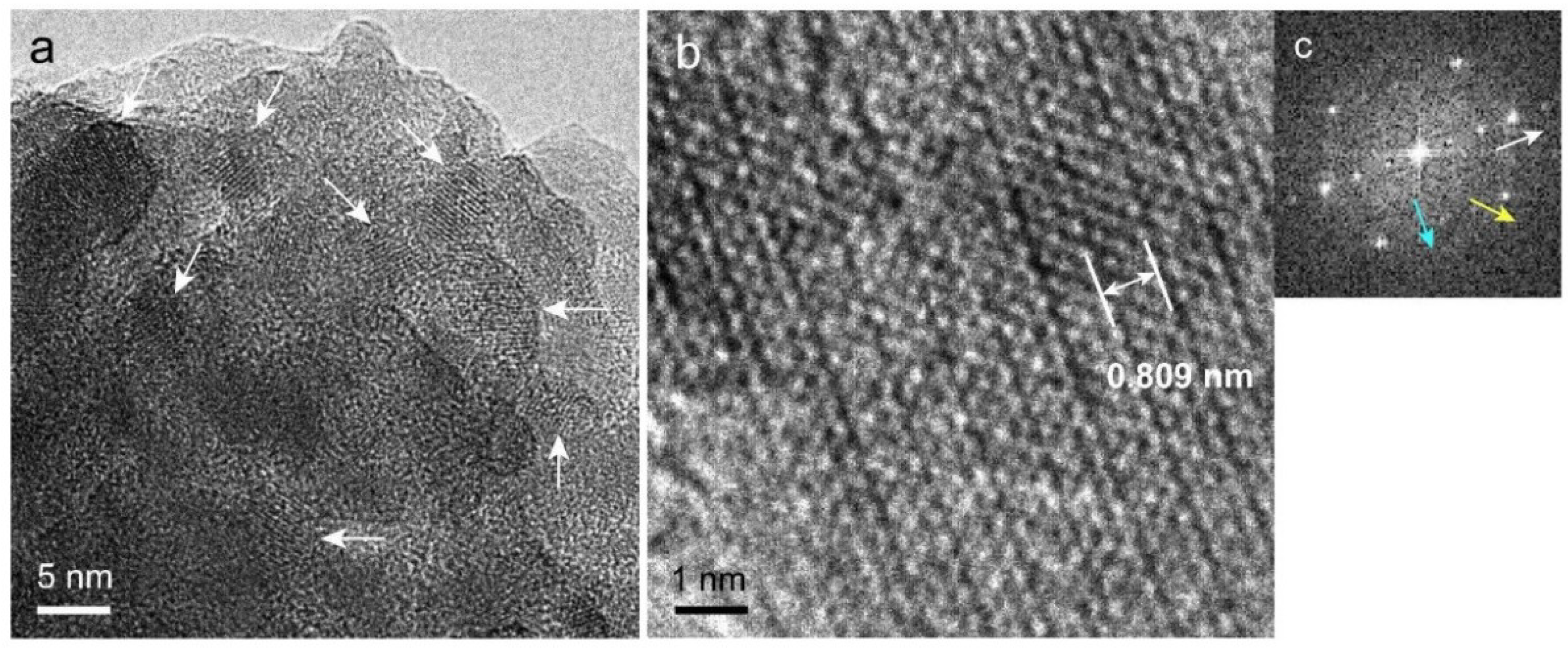
2.7. Transmission Electron Microscopy (TEM) Observation and Selected-Area Electron Diffraction (SAED) Measurement of Precipitated Nodules in a Wide Area
2.8. High-Resolution TEM (HR-TEM) Observation and Fast Fourier Transform (FFT) Image Analysis of Precipitated Nodules in a Narrow Area
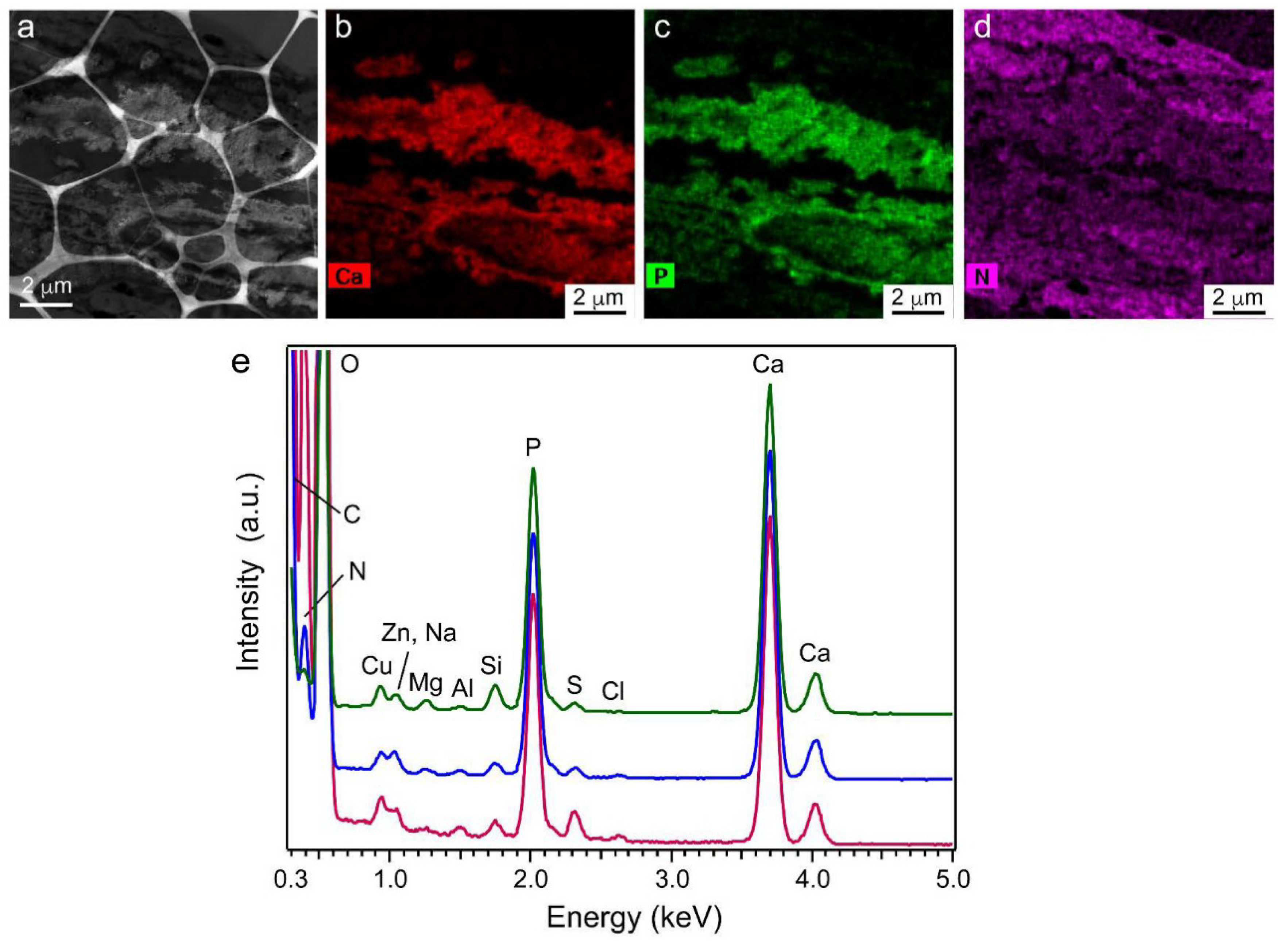
2.9. Element Content in Precipitated Nodules Measured Using Scanning TEM Energy-Dispersive X-ray Spectroscopy (STEM-EDS)
3. Discussions
4. Materials and Methods
4.1. Alkaline Phosphatase (ALP) Activity Assay of the PPU-7 Cell Line
4.2. Alkaline Phosphatase Staining
4.3. Gene Expression of the PPU-7 Cell Line
4.4. Detection of Precipitated Nodules in PPU-7 Cells
4.5. Statistical Analysis
4.6. Characterization of Proteins in the Precipitated Nodules from PPU-7 Cells
4.7. Degree of Supersaturation with Respect to Each Calcium Phosphate Phase
4.8. XRD
4.9. SEM
4.10. TEM Observation and Elemental Analysis Using STEM-EDS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| mRNA | messenger ribonucleic acid |
| SDS | sodium dodecyl sulfate |
| kDa | kilodalton |
| Tris | 2-amino-2-hydroxymethyl-propane-1,3-diol |
| PMSF | phenylmethylsulfonyl fluoride |
| PBS | phosphate buffered saline |
References
- Zafar, M.S.; Khurshid, Z.; Almas, K. Oral tissue engineering progress and challenges. Tissue Eng. Regen. Med. 2015, 12, 387–397. [Google Scholar] [CrossRef]
- Hu, L.; Gao, Z.; Xu, J.; Zhu, Z.; Fan, Z.; Zhang, C.; Wang, J.; Wang, S. Decellularized Swine Dental Pulp as a Bioscaffold for Pulp Regeneration. BioMed Res. Int. 2017, 2017, 9342714. [Google Scholar] [CrossRef] [PubMed]
- Yasui, T.; Mabuchi, Y.; Morikawa, S.; Onizawa, K.; Akazawa, C.; Nakagawa, T.; Okano, H.; Matsuzaki, Y. Isolation of dental pulp stem cells with high osteogenic potential. Inflamm. Regen. 2017, 37, 8. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Imai, M.; Goto, Y.; Murakami, N. Pathological mineralization in a serially passaged cell line from rat pulp. J. Oral Pathol. Med. 1993, 22, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Nagata, T.; Yokota, M.; Ohishi, K.; Nishikawa, S.; Shinohara, H.; Wakano, Y.; Ishida, H. 1α,25-dihydroxyvitamin D3 stimulation of osteopontin expression in rat clonal dental pulp cells. Arch. Oral Biol. 1994, 39, 775–782. [Google Scholar] [CrossRef]
- MacDougall, M.; Selden, J.K.; Nydegger, J.R.; Carnes, D.L. Immortalized mouse odontoblast cell line MO6-G3 application for in vitro biocompatibility testing. Am. J. Dent. 1998, 11, S11–S16. [Google Scholar] [PubMed]
- Thonemann, B.; Schmalz, G. Bovine dental papilla-derived cells immortalized with HPV 18 E6/E7. Eur. J. Oral Sci. 2000, 108, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Iwata, T.; Yamakoshi, Y.; Simmer, J.P.; Ishikawa, I.; Hu, J.C. Establishment of porcine pulp-derived cell lines and expression of recombinant dentin sialoprotein and recombinant dentin matrix protein-1. Eur. J. Oral Sci. 2007, 115, 48–56. [Google Scholar] [CrossRef]
- Kamata, N.; Fujimoto, R.; Tomonari, M.; Taki, M.; Nagayama, M.; Yasumoto, S. Immortalization of human dental papilla, dental pulp, periodontal ligament cells and gingival fibroblasts by telomerase reverse transcriptase. J. Oral Pathol. Med. 2004, 33, 417–423. [Google Scholar] [CrossRef]
- Galler, K.M.; Schweikl, H.; Thonemann, B.; D’Souza, R.N.; Schmalz, G. Human pulp-derived cells immortalized with Simian Virus 40 T-antigen. Eur. J. Oral Sci. 2006, 114, 138–146. [Google Scholar] [CrossRef]
- Yang, X.; van der Kraan, P.M.; van den Dolder, J.; Walboomers, X.F.; Bian, Z.; Fan, M.; Jansen, J.A. STRO-1 selected rat dental pulp stem cells transfected with adenoviral-mediated human bone morphogenetic protein 2 gene show enhanced odontogenic differentiation. Tissue Eng. 2007, 13, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Ohazama, A.; Tucker, A.; Sharpe, P.T. Organized tooth-specific cellular differentiation stimulated by BMP4. J. Dent. Res. 2005, 84, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Shimabukuro, Y.; Ueda, M.; Ozasa, M.; Anzai, J.; Takedachi, M.; Yanagita, M.; Ito, M.; Hashikawa, T.; Yamada, S.; Murakami, S. Fibroblast growth factor-2 regulates the cell function of human dental pulp cells. J. Endod. 2009, 35, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Tjaderhane, L.; Koivumaki, S.; Paakkonen, V.; Ilvesaro, J.; Soini, Y.; Salo, T.; Metsikko, K.; Tuukkanen, J. Polarity of mature human odontoblasts. J. Dent. Res. 2013, 92, 1011–1016. [Google Scholar] [CrossRef]
- Li, Y.; Lu, X.; Sun, X.; Bai, S.; Li, S.; Shi, J. Odontoblast-like cell differentiation and dentin formation induced with TGF-β1. Arch. Oral Biol. 2011, 56, 1221–1229. [Google Scholar] [CrossRef]
- Neves, V.C.; Babb, R.; Chandrasekaran, D.; Sharpe, P.T. Promotion of natural tooth repair by small molecule GSK3 antagonists. Sci. Rep. 2017, 7, 39654. [Google Scholar] [CrossRef]
- Olkkola, K.T.; Ahonen, J. Midazolam and other benzodiazepines. In Modern Anesthetics. Handbook of Experimental Pharmacology; Schuttler, J., Schwilden, H., Eds.; Springer: Berlin, Germany, 2008; Volume 182, pp. 335–360. [Google Scholar]
- Smith, R.; Brown, J. Midazolam for status epilepticus. Aust. Prescr. 2017, 40, 23–25. [Google Scholar] [CrossRef]
- So, E.C.; Chen, Y.C.; Wang, S.C.; Wu, C.C.; Huang, M.C.; Lai, M.S.; Pan, B.S.; Kang, F.C.; Huang, B.M. Midazolam regulated caspase pathway, endoplasmic reticulum stress, autophagy, and cell cycle to induce apoptosis in MA-10 mouse Leydig tumor cells. OncoTargets Ther. 2016, 9, 2519–2533. [Google Scholar]
- Ohno, S.; Kobayashi, K.; Uchida, S.; Amano, O.; Sakagami, H.; Nagasaka, H. Cytotoxicity and type of cell death induced by midazolam in human oral normal and tumor cells. Anticancer Res. 2012, 32, 4737–4747. [Google Scholar] [PubMed]
- Stevens, M.F.; Werdehausen, R.; Gaza, N.; Hermanns, H.; Kremer, D.; Bauer, I.; Kury, P.; Hollmann, M.W.; Braun, S. Midazolam activates the intrinsic pathway of apoptosis independent of benzodiazepine and death receptor signaling. Reg. Anesth. Pain Med. 2011, 36, 343–349. [Google Scholar] [CrossRef]
- Mishra, S.K.; Kang, J.H.; Lee, C.W.; Oh, S.H.; Ryu, J.S.; Bae, Y.S.; Kim, H.M. Midazolam induces cellular apoptosis in human cancer cells and inhibits tumor growth in xenograft mice. Mol. Cells 2013, 36, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shao, H.; Xu, K.Q.; Kuang, L.T.; Chen, R.F.; Xiu, H.H. Midazolam suppresses osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1411–1418. [Google Scholar] [PubMed]
- Sloan, A.J.; Rutherford, R.B.; Smith, A.J. Stimulation of the rat dentine-pulp complex by bone morphogenetic protein-7 in vitro. Arch. Oral Biol. 2000, 45, 173–177. [Google Scholar] [CrossRef]
- Chen, S.; Gu, T.T.; Sreenath, T.; Kulkarni, A.B.; Karsenty, G.; MacDougall, M. Spatial expression of Cbfa1/Runx2 isoforms in teeth and characterization of binding sites in the dspp gene. Connect. Tissue Res. 2002, 43, 338–344. [Google Scholar] [CrossRef] [PubMed]
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Yang, F.; Deng, R.; Li, D.; Song, Z.; Tian, Y.; Wang, R.; Ling, J.; Lin, Z. Smad 1/5 is involved in bone morphogenetic protein-2-induced odontoblastic differentiation in human dental pulp cells. J. Endod. 2012, 38, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, X.; Xu, X.; Mayo, J.; Bringas, P., Jr.; Jiang, R.; Wang, S.; Chai, Y. SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis. Development 2011, 138, 1977–1989. [Google Scholar] [CrossRef]
- Cope, D.W.; Hughes, S.W.; Crunelli, V. GABAA receptor-mediated tonic inhibition in thalamic neurons. J. Neurosci. 2005, 25, 11553–11563. [Google Scholar] [CrossRef]
- Bormann, J. The ‘ABC’ of GABA receptors. Trends Pharmacol. Sci. 2000, 21, 16–19. [Google Scholar] [CrossRef]
- Kim, J.H.; Jeon, M.; Song, J.S.; Lee, J.H.; Choi, B.J.; Jung, H.S.; Moon, S.J.; DenBesten, P.K.; Kim, S.O. Distinctive genetic activity pattern of the human dental pulp between deciduous and permanent teeth. PLoS ONE 2014, 9, e102893. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.; Shin, Y.; Lee, H.S.; Jeon, M.; Kim, S.O.; Cho, S.W.; Ruparel, N.B.; Song, J.S. Comparative Gene Expression Analysis of the Coronal Pulp and Apical Pulp Complex in Human Immature Teeth. J. Endod. 2016, 42, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Potdar, P.D.; Jethmalani, Y.D. Human dental pulp stem cells: Applications in future regenerative medicine. World J. Stem Cells 2015, 7, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Gronthos, S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J. Bone Miner. Res. 2003, 18, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, R.; Oida, S.; Yamakoshi, Y. Dentin Sialophosphoprotein-derived Proteins in the Dental Pulp. J. Dent. Res. 2015, 94, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Niwa, T.; Yamakoshi, Y.; Yamazaki, H.; Karakida, T.; Chiba, R.; Hu, J.C.; Nagano, T.; Yamamoto, R.; Simmer, J.P.; Margolis, H.C.; et al. The dynamics of TGF-β in dental pulp, odontoblasts and dentin. Sci. Rep. 2018, 8, 4450. [Google Scholar] [CrossRef]
- Puchtler, H.; Meloan, S.N.; Terry, M.S. On the history and mechanism of alizarin and alizarin red S stains for calcium. J. Histochem. Cytochem. 1969, 17, 110–124. [Google Scholar] [CrossRef]
- Puchtler, H.; Meloan, S.N. Demonstration of phosphates in calcium deposits: a modification of von Kossa’s reaction. Histochemistry 1978, 56, 177–185. [Google Scholar] [CrossRef]
- Saito, T.; Toyooka, H.; Ito, S.; Crenshaw, M.A. In vitro study of remineralization of dentin: effects of ions on mineral induction by decalcified dentin matrix. Caries Res. 2003, 37, 445–449. [Google Scholar] [CrossRef]
- Yamakoshi, Y.; Lu, Y.; Hu, J.C.; Kim, J.W.; Iwata, T.; Kobayashi, K.; Nagano, T.; Yamakoshi, F.; Hu, Y.; Fukae, M.; et al. Porcine dentin sialophosphoprotein: length polymorphisms, glycosylation, phosphorylation, and stability. J. Biol. Chem. 2008, 283, 14835–14844. [Google Scholar] [CrossRef]
- Onuma, K.; Iijima, M. Artificial enamel induced by phase transformation of amorphous nanoparticles. Sci. Rep. 2017, 7, 2711. [Google Scholar] [CrossRef]
- Teruel Jde, D.; Alcolea, A.; Hernandez, A.; Ruiz, A.J. Comparison of chemical composition of enamel and dentine in human, bovine, porcine and ovine teeth. Arch. Oral Biol. 2015, 60, 768–775. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Oida, S.; Suzuki, S.; Iwata, T.; Yamakoshi, Y.; Ogata, Y.; Gomi, K.; Arai, T.; Fukae, M. Porcine Enamel Protein Fractions Contain Transforming Growth Factor-β1. J. Periodontol. 2006, 77, 1688–1694. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Nancollas, G.H. Calcium orthophosphates: crystallization and dissolution. Chem. Rev. 2008, 108, 4628–4669. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Esteban, R. A Drug-Centric View of Drug Development: How Drugs Spread from Disease to Disease. PLoS Comput. Biol. 2016, 12, e1004852. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.S.; Patel, C.J. A standard database for drug repositioning. Sci. Data 2017, 4, 170029. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karakida, T.; Onuma, K.; Saito, M.M.; Yamamoto, R.; Chiba, T.; Chiba, R.; Hidaka, Y.; Fujii-Abe, K.; Kawahara, H.; Yamakoshi, Y. Potential for Drug Repositioning of Midazolam for Dentin Regeneration. Int. J. Mol. Sci. 2019, 20, 670. https://doi.org/10.3390/ijms20030670
Karakida T, Onuma K, Saito MM, Yamamoto R, Chiba T, Chiba R, Hidaka Y, Fujii-Abe K, Kawahara H, Yamakoshi Y. Potential for Drug Repositioning of Midazolam for Dentin Regeneration. International Journal of Molecular Sciences. 2019; 20(3):670. https://doi.org/10.3390/ijms20030670
Chicago/Turabian StyleKarakida, Takeo, Kazuo Onuma, Mari M. Saito, Ryuji Yamamoto, Toshie Chiba, Risako Chiba, Yukihiko Hidaka, Keiko Fujii-Abe, Hiroshi Kawahara, and Yasuo Yamakoshi. 2019. "Potential for Drug Repositioning of Midazolam for Dentin Regeneration" International Journal of Molecular Sciences 20, no. 3: 670. https://doi.org/10.3390/ijms20030670
APA StyleKarakida, T., Onuma, K., Saito, M. M., Yamamoto, R., Chiba, T., Chiba, R., Hidaka, Y., Fujii-Abe, K., Kawahara, H., & Yamakoshi, Y. (2019). Potential for Drug Repositioning of Midazolam for Dentin Regeneration. International Journal of Molecular Sciences, 20(3), 670. https://doi.org/10.3390/ijms20030670