Abstract
A PACLOBUTRAZOL-RESISTANCE (PRE) gene family, consisting of six genes in Arabidopsis thaliana, encodes a group of helix-loop-helix proteins that act in the growth-promoting transcriptional network. To delineate the specific role of each of the PRE genes in organ growth, we took a reverse genetic approach by constructing high order pre loss-of-function mutants of Arabidopsis thaliana. In addition to dwarf vegetative growth, some double or high order pre mutants exhibited defective floral development, resulting in reduced fertility. While pre2pre5 is normally fertile, both pre2pre6 and pre5pre6 showed reduced fertility. Further, the reduced fertility was exacerbated in the pre2pre5pre6 mutant, indicative of the redundant and critical roles of these PREs. Self-pollination assay and scanning electron microscopy analysis showed that the sterility of pre2pre5pre6 was mainly ascribed to the reduced cell elongation of anther filament, limiting access of pollens to stigma. We found that the expression of a subset of flower-development related genes including ARGOS, IAA19, ACS8, and MYB24 was downregulated in the pre2pre5pre6 flowers. Given these results, we propose that PREs, with unequal functional redundancy, take part in the coordinated growth of floral organs, contributing to successful autogamous reproduction in Arabidopsis thaliana.
1. Introduction
Coordinated organ growth is essential for the determination of organ architecture and its biological functions. The process of pollination in autogamous plants is a good example to study how proper organ growth is critical for the function of organs. For successful pollination, the coordinated growth of anther filament and style, as well as the release of functional pollens, is essential. Thus filament growth and anthesis must be tightly controlled and keep pace with the pistil development for successful fertilization. Indeed, dysregulated growth of these floral organs such as shortened anther filament or longer style, termed as heterostyly, results in allogamous reproduction, requiring cross-pollination [,,].
Organ growth is driven by cell division and elongation, which are controlled by integrated signals from environments and the endogenous development program mediated by plant hormones. For example, the stamen development is controlled by multiple phytohormones including GA, BLs, auxins, and JA [,,,]. Deficiency of those hormones causes male sterility [,,,,,,,,]. The hormonal signals are either converged or independently transduced to a transcriptional regulatory network, involving transcription factors such as MYB21/MYB24 and ARF6/ARF8. In support, the mutants of myb21myb24 or arf6arf8 display severe sterility, primarily due to reduced floral organ growth [,,]. The expression of MYB21/MYB24 is regulated by multiple plant growth hormones including GA, JA, and auxin in part with a hierarchical relationship. GA and ARF6/ARF8 regulate JA-biosynthetic genes to regulate the production of JA, which in turn activates the expression of MYB21/MYB24 [,].
A small gene family, designated as PREs (PACLOBUTRAZOL-RESISTANCE), encode atypical helix-loop-helix (HLH) proteins in higher plants. In Arabidopsis thaliana, PRE family comprise six genes, including PRE1/BNQ1(BANQUO1), PRE2/BNQ2, PRE3/BNQ4/ATBS1(ACTIVATION-TAGGED bri1 SUPPRESSOR1)/TMO7 (TARGET OF MP7), PRE4/BNQ3, PRE5, PRE6/KDR (KIDARI) [,,,,,]. Since PRE1 has been initially characterized as a positive regulator of GA signaling [], subsequent studies have elucidated that PREs are also involved in growth regulation by a wide range of signals, including BR, auxin, high temperature and light [,,,,,]. Elevated expression of any single PRE gene confers elongated hypocotyls/petioles, while downregulation of multiple PRE genes by gene silencing caused dwarf phenotypes, suggestive of functionally redundant roles of PREs to promote cell elongation. How PRE functions in cell elongation has been revealed by interactome studies [,,,,]. Yeast two-hybrid screening of PRE interacting proteins has identified distinct classes of atypical HLH proteins such as IBH1/AIFs, PAR1/PAR2, and HFR1. These atypical HLH proteins were shown to be capable of inhibiting cell elongation, presumably through heterodimerization with bHLH proteins with growth-promoting activity, including HBI1/ACEs, BEEs or PIFs, respectively. Based on these results, it has been postulated that PRE functions in the PRE-IBH1-HBI1/ACEs or PRE-PAR1/PAR2/HFR1-PIF tripartite HLH/bHLH modules for transcriptional reprogramming, resulting in cell elongation.
Most of the functional studies of PRE genes have been based on the transgenic plants that ectopically express each of the PRE genes or transgenic plants with gene silencing of multiple PREs, hampering the assessment of the function of individual PREs [,,,,]. Thus, the in vivo function of individual PRE gene member remains to be determined. Despite functional similarity or redundancy among PRE genes, the affinity toward its interacting proteins appeared to be differential [,]. Besides, expression analysis of the differential inducibility of PRE genes by GA, auxin, or BR as well as differential tissue-preferential expression pattern implicated distinct roles of PRE genes [,,,]. Indeed, under a specific environmental condition or developmental context, a single pre mutant could exhibit developmental alterations [,,]. However, redundant roles among PREs still remain to be dissected through systematic high order loss-of-function mutant analysis.
In the present study, we generated multiple combinations of loss-of-function pre mutants and investigated the defect in organ growth in Arabidopsis thaliana. While single and a subset of double pre mutants did not show discernible growth defects, some combination of high order pre mutants showed shortened hypocotyls/petioles, recapitulating the known role of PREs during vegetative organ growth. Further, we also found that certain combination of pre mutations, including pre2pre6, pre5pre6, and pre2pre5pre6 caused shortened anther filaments, resulting in failure in pollination. Microscopic analysis and gene expression analysis indicated that the reduced fertility of high-order pre mutants is due to reduced cell elongation of anther filament, which is accompanied by altered transcriptional expression of several flower-development related genes, including ARGOS, ACS8, IAA9, and MYB24. By comparing phenotypes of different combinations of high order pre mutants, we suggest that PRE gene family members take part, in an unequally redundant manner, in the regulation of cell elongation/expansion for diverse aspects of growth and development.
2. Results
2.1. Unequally Redundant Role of PREs during Vegetative Organ Growth
A small gene family, designated as PRE (PACLOBUTRAZOL RESISTANCE), has been characterized based on gain-of-function studies [,,,,,]. Functional redundancy among PRE genes was inferred by the findings that the transgenic plants that overexpressed any single PRE member displayed similar developmental alterations. In support of the functional similarity among PRE members, we found that all of the PRE members in Arabidopsis thaliana could interact with PAR1 or PAR2, a class of PRE-interacting HLH proteins [], in yeast two-hybrid assay (Figure S1).
Based on differential spatiotemporal and hormone-responsive expression patterns of PREs (Figure S2) [,,,], we hypothesized that each PRE protein plays a specified, if redundant, role during various stages of growth and development, which might constitute an evolutionary selection pressure to preserve multiple, functionally redundant [,,], PRE genes. Indeed, a few SNP/deletion/insertion variations with a high impact were present in the PRE genes among 1,135 natural accessions of Arabidopsis thaliana (Table S1) []. To elucidate the function of PRE gene members in Arabidopsis thaliana, we obtained T-DNA insertional mutants of each PRE gene from The Arabidopsis Information Resource database (TAIR, www.arabidopsis.org). By RT-PCR analysis, we found that mutants of pre2, pre5, or pre6 were a null allele, while the mutant of PRE4 was a hypomorphic allele, though it harbors T-DNA in its intron (Figure S3). The pre1 mutant was not studied further since we found that a PRE1 T-DNA mutant (GK646C02) contains a T-DNA insertion in the 3′-UTR region, which did not alter the transcript level. For the PRE3 gene, there is no available T-DNA insertional mutant for the present.
To dissect the functional redundancy among PRE genes, we systematically generated a series of high order pre mutants including double mutants of pre2pre5, pre2pre6, pre2pre4, pre4pre5, pre4pre6, or pre5pre6, triple mutants of pre2pre4pre6, pre2pre5pre6, or pre4pre5pre6 and quadruple mutant of pre2pre4pre5pre6. In the course of generating the high order double mutant series, we observed several developmental abnormalities in certain combinations of pre mutations, revealing unequal redundancy among PRE genes. In agreement with the results of RNAi-based silencing of multiple PREs [,], high order pre mutants, e.g., pre2pre5pre6 exhibited a dwarf growth phenotype and reduced hypocotyl elongation under light condition (Figure 1 and Figure S4). Direct comparison of the leaf development of single, double, triple and quadruple pre mutants showed that PRE2 and PRE6 play dominant roles during vegetative growth. As shown in Figure 1, petiole growth was significantly reduced in the pre2pre6 mutant. On the contrary, the shortened petiole was not observed or marginally changed in other double mutant series, including pre2pre4, pre2pre5, pre4pre5, pre4pre6 and pre5pre6, and a triple mutant, pre4pre5pre6. Notably, the dwarf growth of pre2pre6 was not exacerbated in the triple mutants of pre2pre4pre6, pre2pre5pre6. The dwarf growth of pre2pre5pre6 was also comparable to that of the quadruple mutant, pre2pre4pre5pre6 (Figure 1A). The leaf length, but not leaf width, was also significantly reduced in the pre2pre6 double mutant and triple mutants, including pre2pre6, pre2pre4pre6 and pre2pre5pre6, resulting in orbicular leaf shape (Figure 1A,C). These results implicate the dominant roles of PRE2 and PRE6 during leaf development. The scanning electron microscopy analysis showed that the cell length was clearly reduced in the petioles of pre2pre5pre6 compared to wild type, suggesting that the shortened petiole phenotype is mainly due to reduced cell elongation (Figure 1D).
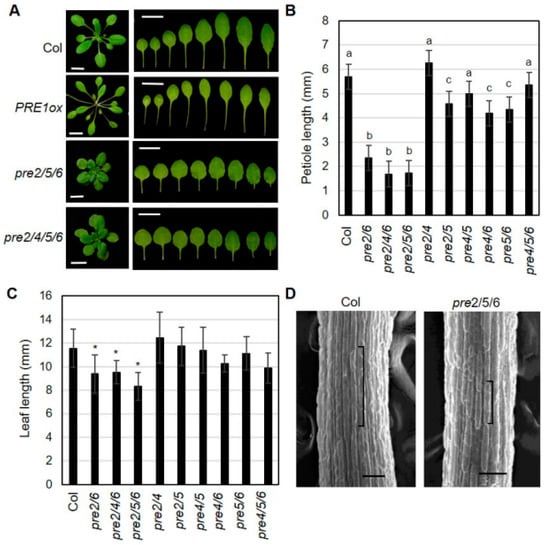
Figure 1.
High order loss-of-function of PREs alters leaf growth. (A) Representative leaf morphology of pre mutants. The pictures were taken from 3 week-old plants. The leaves were arranged according to the age from left to right. Scale bar, 1 cm. (B) Petiole length of pre mutants. The petiole length was measured with the 7th leaves of 3 week-old plants grown under long-day (16h light/8h dark). Values are mean ± SD (n = 12–15). Significant difference among wild type (Col) and mutants are indicated with letters (ANOVA, Tukey HSD test, p < 0.05). (C) Leaf length was analyzed from the same leaves as described in B. Values are mean ± SD (n = 12–15). A significant difference between wild type (Col) and mutants is indicated with asterisks (t-test, * p < 0.01). (D) Cell length of petiole epidermis. Petioles of the third leaf of wildtype (Col) or pre2pre5pre6 were subject to scanning electron microscopy. Brackets indicate the lengths of single epidermal cells from the middle of petioles. Scale bar, 100 μm.
As PRE genes have been implicated in the hypocotyl elongation, we also examined hypocotyl growth of high order pre mutant series. As shown in Figure S4, pre2pre4pre6, pre2pre5pre6 and pre2pre4pre5pre6, but not pre4pre5pre6, showed shortened hypocotyl growth under light condition. Among the double mutant series, only pre2pre6 displayed significantly shorter hypocotyl phenotype, compared to wild type, which is in line with the prominent role of PRE2 and PRE6 in the petiole elongation. However, we could not see any noticeable defects in the dark-grown seedlings, even in the pre2pre4pre5pre6 quadruple mutant (Figure S4). Unlike the aerial part of vegetative organs, high order pre mutants exhibited normal root growth phenotype as well as normal flowering phenotypes, while PRE1 overexpressing mutants displayed longer root length and early-flowering phenotypes, compared to wild type plant (Figure S5). These results implicate the function of other PRE members, PRE1 and PRE3, may still support normal root growth and flowering phenotypes.
2.2. Unequally Redundant Role of PREs during Flower Maturation
In addition to vegetative organ growth, we observed defects of reproductive organ growth in specific combinations of pre mutants. While other single pre mutant did not show discernible differences in flowering and seed setting, a few early formed flowers in pre6 failed to set seeds (Figure S3). Further, the weak sterility of pre6 was dramatically exaggerated by additional loss-of-function of PRE2 or PRE5 genes (Figure 2A). The double mutants of pre2pre6 and pre5pre6 showed fertility of about 35% of fully mature seeds, whereas the fertility of pre4pre6 was similar to that of pre6. In contrast, pre2pre4, pre2pre5, or pre4pre5 mutants showed normal fertility, compared to wild type. These results implied that PRE6 plays a critical role, redundantly with PRE2 or PRE5, in the reproductive process. Supporting this, the pre2pre5pre6 triple mutant showed very low fertility such that it was difficult to get any mature siliques. In the case of other triple mutants, the fecundity of pre2pre4pre6 or pre4pre5pre6 was comparable to that of pre2pre6 or pre5pre6 double mutants (Figure 2A), which is indicative of the marginal effect of the pre4 mutation.
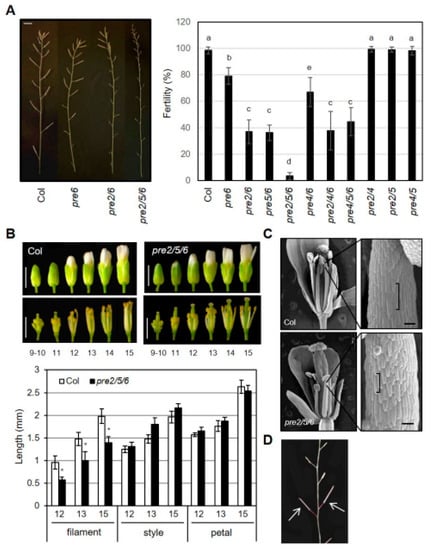
Figure 2.
High order loss-of-function PREs causes sterility. (A) Fertility of pre mutants. Primary inflorescence (left) and fertility of pre mutants (right). The fertility was measured as the percentage of mature siliques per whole siliques of the primary inflorescence. Values are mean ± SD (n = 13–15). Significant difference among wildtype (Col) and mutants are indicated with letters (ANOVA, Tukey HSD test, p < 0.01). Scale bar, 5 mm. (B) Floral organ growth of wildtype (Col) and pre2pre5pre6 during flower development. Upper, Representative flower morphology at indicated flower developmental stages. Scale bar, 1 mm. Lower, The length of the filament, style, petal was measured at indicated flower developmental stages. Values are mean ± SD (n = 15). Asterisk indicates significant difference from wild type (t-test, * p < 0.01). (C) Reduced cell size in pre2pre5pre6 anther filament. Scanning electron microscopy was performed on anther filament from flowers at stage13. Brackets indicate the lengths of single epidermal cells of anther filaments. Scale bar, 100 μm. (D) Restored fertility of pre2pre5pre6 by self-pollination. The pre2pre5pre6 flowers which were manually pollinated with their pollens produced fully mature siliques, as indicated by arrows.
To verify that the loss of PRE genes was responsible for the sterile phenotypes, we performed transgenic complementation by overexpressing PRE6 or PRE2 in the pre2pre6 mutant background. The results showed that multiplmente independent lines of either PRE6 or PRE2 overexpression restored the reduced fertility of pre2pre6 plants, demonstrating that the loss of function of these PREs is responsible for the reduced fertility in the double mutant (Figure 3 and Figure S6).
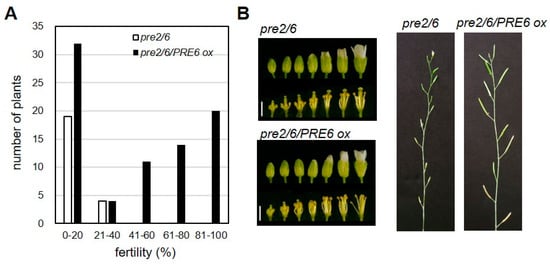
Figure 3.
PRE6 overexpression rescued the sterility of pre2pre6. The pre2pre6 plants were transformed with Agrobacterium carrying the PRE6 overexpressing vector. Full-length cDNA of PRE6 was cloned into the plant overexpression vector. The primary transformants (T1) were selected and subjected to fertility analysis. (A) Fertility of T1 plants was scored by percentage of fully mature siliques per total flowers of the primary inflorescence. We analyzed 81 T1 plants of overexpressing PRE6 in pre2pre6 mutant background. (B) Left, representative flower morphology at various developmental stages. Scale bar, 1 mm. Right, representative primary inflorescences of pre2pre6 and a transgenic line overexpressing PRE6.
Next, we examined the development of floral organs of the pre2pre5pre6 mutant in detail to understand the cellular basis of the reduced fertility. The morphology of bud and style was indistinguishable between wild type and the triple mutant at various flower developmental stages (Figure 2B). On the other hand, the length of anther filament in the pre2pre5pre6 was clearly shorter than that in wild type, suggesting that the reduced elongation of anther filament in the pre2pre5pre6 might limit access of its pollens to stigma. Scanning electron microscopy (SEM) analysis revealed that the shorter anther filament of the pre2pre5pre6 is mainly due to the reduced cell length of anther filament (Figure 2C). To test whether the pollens of the pre2pre5pre6 mutant are functional for fertilization, we performed self-pollination assay with the pre2pre5pre6 mutant by manual crossing. The self-pollinated mutant flowers could set fully mature seeds (Figure 2D), indicating that the reduced fertility in the pre2pre5pre6 mutants may not be due to defective pollen development or anther dehiscence.
The sterility and floral phenotype of the pre2pre5pre6 mutant were reminiscent of defects seen in mutants of transcriptional regulators, MYB21 or ARF6/ARF8 [,], which impair proper growth of anther filament. Previous expression analysis has suggested that PRE genes are auxin-inducible, being targeted by auxin-response factors including ARF5/ARF6/ARF8 [,,,]. In agreement, our RT-PCR analysis indicated that PRE1, PRE2, PRE5, and PRE6 were downregulated in the arf6arf8 double mutant or myb21 mutant (Figure S7). Keeping in mind the hypothesis that PREs would act as a transcriptional regulator, we questioned whether the expression of those flower development-related genes or their target genes were altered in the pre mutant flowers. To address this question, we analyzed the expression of a selected set of genes including MYB24, MYB21, ARF6, ARF8, ARGOS, IAA19, and ACS8 in the flowers at various developmental stages. The results showed that expression of MYB24, ARGOS, IAA19, and ACS8 was downregulated in the pre2pre5pre6 at stage 12 and 13, compared to wild type. In contrast, the transcript level of MYB21 and ARF6 was not altered in the pre2pre5pre6. In the case of ARF8 expression, it was rather upregulated in the pre2pre5pre6, compared to wild type in the floral primordia and flower at stage 15 (Figure 4). Thus, these results implied that these PREs are involved in regulation of a subset of flower-developmental genes such as ARGOS, ACS8, IAA19, and MYB24 acting presumably at the downstream of ARF6/ARF8-MYB21/MYB24 network.
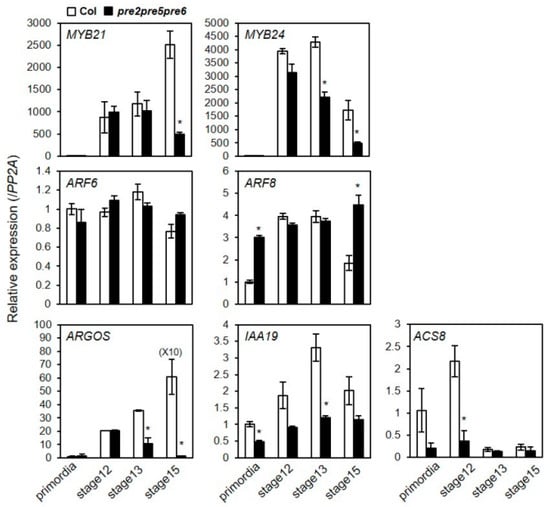
Figure 4.
The expression of flower development-related genes was downregulated in pre2pre5pre6 flowers. Total RNAs were extracted from flowers at indicated flower developmental stages and relative transcript levels were examined by quantitative RT-PCR analysis. The PP2A gene was analyzed as an internal control. Values are mean ± SD from three technical replicates. Significant difference between wild type (Col) and pre2pre5pre6 is indicated with asterisk (t-test, * p < 0.01).
2.3. Expression of PRE6 during Flower Development
We performed real-time qRT-PCR analysis to investigate the spatiotemporal expression pattern of PREs during flower development in detail. The results showed that the expression of most of PREs, except PRE2, appear to correlate with the organ growth phase (Figure 5A). In the case of PRE2, its expression level was shown to be highest in the flower primordia, implying that PRE2 might act at the very early stage of flower maturation. Given the prominent role of PRE6 in the anther filament growth, we generated PRE6pro: GUS transgenic plants to analyze the spatial expression pattern of PRE6 during flower development. The results of GUS staining showed that PRE6 is expressed in various floral tissues, including sepals, a specific part of the style, just below the stigma, and the anther filament (Figure 5B). The expression of PRE6 in anther filaments increased as filaments elongated, in accordance with the role of PRE6 in cell elongation (Figure 5B). During vegetative growth, qRT-PCR analysis combined with promoter-GUS analysis indicated that the expression of PRE6 increased in the actively growing, young leaves, while declined in mature leaves (Figure S8). Together, these results implied that the expression pattern of PRE6 coincides largely with its functional aspects.
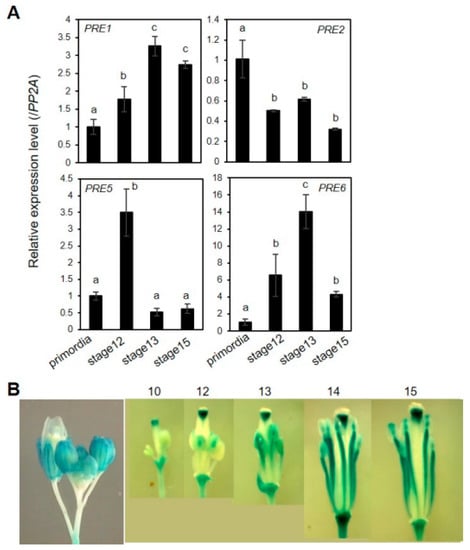
Figure 5.
Expression of PRE genes during flower maturation. (A) Quantitative RT–PCR analyses of gene expression of PREs in developing flowers. Total RNAs were isolated from flowers at specific flower developmental stages. The data was represented as a relative value to primordia that was set to 1. The PP2A gene was used as an internal control. Values are mean ± SD from three technical replicates. Significant difference among flower development stages is indicated with letters (ANOVA, Tukey HSD test, p < 0.05). (B) PRE6pro: GUS analysis. PRE6 promoter activity was monitored by GUS staining of transgenic plants expressing PRE6pro: GUS at indicated flower developmental stages.
3. Discussion
The presence of a functionally redundant PRE gene family, comprised of six genes in Arabidopsis thaliana, raised an intriguing question about the in planta role of individual PRE genes during development. Although ectopic expression of each PRE gene induces very similar developmental alterations, including increased germination potential, early flowering, and elongated vegetative organ growth [], the differential spatial expression pattern implied that PREs might play differential functions depending on the developmental contexts (Figure S2) []. Further, it has been well documented that expression of PRE genes showed differential inducibility upon various plant hormones or environmental signals [,].
In this study, we tried to identify the organ-specific role of PREs by performing reverse genetic analysis with multiple high order loss-of-function mutants of PREs. Our results not only recapitulated the role of PREs during vegetative growth based on artificial microRNA-mediated knock-down of multiple PREs [,], but also revealed their unequally redundant roles for floral organ growth, contributing to successful reproduction (See below). The single pre loss-of-function mutants and most of the double mutants we tested including pre2pre4, pre2pre5, pre4pre5, pre4pre6, and pre5pre6 did not show altered organ growth, which is suggestive of functional compensation by other PREs. The reduced growth of hypocotyl and petiole was evident only in the mutants including both the pre2 and pre6; pre2pre6, pre2pre4pre6, pre2pre5pre6, and pre2pre4pre5pre6. In contrast, the high order pre mutants, pre2pre4, pre2pre5, pre4pre5, pre5pre6, pre4pre6, and pre4pre5pre6 did not exhibit any alteration in vegetative growth. Thus, these results indicated both PRE2 and PRE6 play dominant roles, in a redundant way, for the regulation of vegetative development, including hypocotyl and petiole/leaf growth.
While previous expression analysis showed that PREs are expressed throughout floral development, regulated by floral homeotic genes (AP3/PI) or floral developmental regulators, ARF6/ARF8 [,], functional roles of PREs have not been assessed during flower development. Here, we report the role of PREs as a regulator of anther filament growth. Loss of PRE6 caused slightly reduced fertility, which was exacerbated by the additional loss of PRE2 or PRE5. In support of the roles of PREs in fertility, transgenic expression of PRE2 or PRE6 complement the reduced fertility of the pre2pre6 mutant (Figure 3 and Figure S6). In contrast, pre2pre5 and pre4pre6 did not show reduced fertility phenotype. Thus, it is conceivable that PRE6 plays a dominant role in the regulation of growth of anther filament, partly compensated by PRE2 and PRE5. In agreement with functional redundancy among PRE2, PRE5, and PRE6, the pre2pre5pre6 mutant could rarely set seeds without manual self-crossing. In accordance with the roles of PRE for growth of anther filament, publicly available transcriptome variation analysis showed that these PREs (PRE2, PRE5, and PRE6) are strongly expressed in anther filaments during floral development (Figure S2) []. Since other PREs, such as PRE1 are also expressed in other floral organs, including sepals, petals, and carpels, it is likely that these PREs may be functional to support normal growth in other floral organs in the pre2pre5pre6 mutant. Further studies with higher order pre mutants using a null allele of PRE1, PRE3 or PRE4 will shed light on additional roles in flower maturation.
SEM analysis indicated that the sterility of the pre2pre5pre6 mutant was mainly ascribed to reduced cell elongation of anther filament (Figure 2). However, it is not yet known how PREs control cell elongation of anther filament. Previous microarray analysis of arf6arf8 and myb21 revealed that PRE1 and PRE2 are down-regulated at stage 12 and 13 [,]. This was confirmed by our RT-PCR analysis (Figure S7), suggesting that PREs would act downstream of the ARF6/ARF8-MYB21/MYB24 transcriptional regulatory network for flower maturation. In line with, we found that the expression of ARGOS, IAA19, and ACS8 was significantly reduced in the pre2pre5pre6 at stages when anther filament grows vigorously. As these genes are auxin-inducible and required for flower maturation [,], it is tempting to speculate that the sterility of the pre mutants might entail altered auxin responses and auxin-responsive growth. In line with the hypothesis, PRE has also been implicated in auxin-dependent responses such as hypocotyl growth and root development [,,]. Thus, it is conceivable that a gene regulatory network for flower maturation involves a circuit of transcriptional regulators including ARF-PRE-IBH1-HBI/ACEs, which has been proposed for a transcriptional network for cell elongation during seedling establishment []. In agreement, it has been reported that constitutive overexpression of IBH1 caused smaller flowers, with smaller cell length, while transgenic ACE1-overexpressing plants produced bigger flowers [].
Our results showed that the effect of the pre5 mutation is more prominent in the reproductive stage, while that of pre2 and pre6 was evident during both vegetative growth and reproductive development (Figure 1 and Figure 2). In the case of the pre4 mutant, it marginally affected developmental aspects examined in this study. The absence of phenotypic effects could be due to its hypomorphic nature (Figure S3). Indeed, we did not observe a pale green sepal phenotype in the pre4 mutant, which is a representative visible phenotype of a null allele of PRE4 []. Alternatively, PRE4 may play primary roles in certain environmental and developmental contexts not covered in this study. Considering that PRE4, along with other PREs such as PRE3, is abundantly expressed in the root (Figure S2), it would be interesting to test root growth phenotypes of high order pre mutants under various environmental conditions. Nonetheless, our results suggest that each PRE gene plays an unequally redundant role. For the time being, the molecular basis of the unequal redundancy has not been clearly addressed. In the case of PREs, the expression pattern appears to correlate largely to its biological functions. Several growth-promoting stimuli such as GA, BR, auxin, shade, high temperature were shown to entail upregulation of some, if not all, of PRE genes differentially [,,,,]. In line with these findings, we also observed prominent expression of PRE6 during leaf growth as well as floral organs, including anther filament (Figure 5 and Figure S8). Assuming that PREs function by forming a heterodimer with other HLH/bHLH proteins [,,], it also cannot be ruled out that each PRE might have a differential affinity to its binding proteins. Indeed, IBH1 was reported to interact with PRE1, PRE3, PRE4, and PRE5, but not with PRE2 and PRE6 [], whereas AIF1, another PRE-interacting protein, was shown to interact with PRE3, but not with PRE1 [,]. In contrast, PAR1 and PAR2, another class of PRE-interacting HLH proteins [], could interact with all of the PRE proteins (Figure S1). As such, the differential expression of PRE-interacting proteins might also contribute to functional specificity among PREs. Thus, further study toward systems-wide understanding will determine the molecular basis of unequal redundancy among PRE genes under various environmental and developmental contexts.
4. Materials and Methods
4.1. Plant Materials
Arabidopsis thaliana ecotype Columbia (Col-0) was used as a wildtype in this study. The mutants of myb21-5 (myb21) and arf6-2arf8-3 (arf6arf8) have been previously described [], and were obtained from Dr. J Reed (University of North Carolina-Chapel Hill, USA). T-DNA insertional mutants of PRE genes were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/) and the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/); N122628 (pre2), SALK_098908C (pre4), SALK_013011 (pre5), and SALK_048383 (pre6). To make double pre mutants, the single T-DNA insertional mutants were crossed with each other. Triple and quadruple mutants of pre2pre5pre6, pre2pre4pre6, pre4pre5pre6 and pre2pre4pre5pre6 mutants were generated by crossing of pre2pre5 with pre5pre6 or pre2pre4 with pre5pre6. The genotypes of high order pre mutants were confirmed by genotyping PCR and RT-PCR analysis with gene-specific primer pairs (Table S2).
4.2. Phenotypic Analysis
Unless otherwise stated, seeds were surface sterilized, kept at 4 °C for 3 days, and plated on the half strength of MS medium (Duchefa, Haarlem, Netherlands) supplemented with 0.5% sucrose and 0.7% phytoagar. After 7 d, they were transferred to soil (Sunshine Professional Growing Mix #5) for further growth under long days (16 h light/8 h dark) at 21 to 23 °C. For hypocotyl measurement, seedlings were grown either under short-day (8 h light/16 h dark) W light (15 μW cm−2) or in the dark for 4 days. For measurement of petiole and leaf length, plants were grown under long-day light conditions (16 h light/8 h dark). Petiole and leaf lengths were measured from the 7th leaf of 3 weeks old plants. The floral bud and dissected floral organs at different flower development stages, denoted by Smyth et al. [], were taken and scanned using a flatbed scanner to measure organ lengths. The measurement of organ length was performed using Image J software (https://imagej.nih.gov/ij/). Flowering time was measured by rosette leaf numbers when the first clear appearance of a flower petal that was visible. Fertility was denoted by the percentage of fertile siliques, bearing seeds, among flowers formed on the primary inflorescence at 40 days after growth. For scanning electron microscopy, petioles and flowers were fixed and processed as described []. Briefly, petioles and flowers were fixed with modified Karnovsky’s fixative (2% paraformaldehyde, 2% glutaraldehyde in 0.05 M sodium cacodylate buffer, pH 7.2) for 2 h at 4 °C. After washing with 0.05 M sodium cacodylate buffer (pH 7.2), the sample was fixed further in 1% osmium tetroxide for 2 h at 4 °C. Washed sample was dehydrated with a series of Ethanol from 30% to 100%. After critical point drying (Balzer’s critical point drier), the samples were mounted on aluminum stubs and sputter-coated (SC7620; Quorum) with gold for SEM imaging. The specimens were imaged in a Field-Emission Scanning Electron Microscope (ZEISS, Oberkochen, Germany).
4.3. Transgenic Complementation of pre2pre6
Full-length cDNA of PRE2 or PRE6 was amplified with specific primer sets designed with the overhang of restriction enzyme site, KpnI and XhoI for PRE2 and BamHI and XhoI for PRE6 respectively; 5′-GGCGGTACCCAATGTCTTCTAGCAGAAGG-3′ and 5′-GGCCTCGAGGTTCCATTAATCAAGCTCCT-3′ for PRE2 and 5′- GGATCCAGATGTCTAGCAGAAGATCATC-3′ and 5′-CTCGAGATATAATTAAGCAAGCTCCTAA-3′ for PRE6. The amplified PCR products were subcloned into the pGEM-T easy vector (Promega, Madison, USA). After verification no sequence error, the vector containing PRE2 or PRE6 was digested with KpnI/XhoI or BamHI/XhoI for PRE2 or PRE6, respectively. The digest was gel-eluted and ligated with pENTR1A digested with the same restriction enzyme set (Invitrogen, Carlsbad, CA, USA). The cloned entry clone was recombined into a gateway-compatible plant expression vector, pH2GW7 using LR clonase [], respectively. The resulting construct was transformed into Agrobacterium tumefaciens strain GV3101, which was used for the transformation of the pre2pre6 plants by the floral dipping method []. More than 30 transgenic lines with T-DNA insertions were isolated and used for physiological analysis.
4.4. Expression Analysis
Total RNA from different tissues was extracted with a Spectrum total RNA extraction kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instruction. One µg of RNA was reverse transcribed by using RevertAid™ M-MuLV Reverse Transcriptase (Fermentas, Waltham, MA, USA). The 10-fold diluted cDNA was subject to RT-PCR analysis or qRT-PCR analysis using primers listed in Table S2. The qRT-PCR analysis was performed with an Eco™ Real-Time PCR System (Illumina, San Diego, CA, USA) using a QuantiMix SYBRT kit (Philekorea, Seoul, South Korea). The reactions were performed in triplicate for each run. The relative expression level of each gene was determined after being normalized to that of PP2A (At1g13320). The comparative 2–ΔΔCt method was used to evaluate the relative quantities of each amplified product in the samples, following the procedure as described [].
4.5. Yeast Two-hybrid Analysis
Yeast two-hybrid (Y2H) assays were performed following the procedure as described in []. The PRE cDNAs were cloned into the pGilda vector (Clontech, Mountain View, USA), except for PRE6 which is subcloned to gGilda by LR recombination to generate a LexA-DB fused construct. PRE1-PRE5 cDNA was amplified with specific primer set for pGilda cloning. All primer sets include EcoRI and XhoI restriction enzyme site for cloning into the pGilda vector. The amplified fragments were cloned into a pGEM-T easy vector. After verification of no sequence error, the digested PRE cDNA fragments were ligated with pGilda vector. The full-length of the PAR1 and PAR2 cDNAs were cloned into pYesTrp2 vector (Invitrogen, Carlsbad, CA, USA) to produce the B42-AD fused protein. The specific primer sets for PAR1 and PAR2 include HindIII and XhoI restriction enzyme sites for cloning into the pYesTrp2 vector. After cloning into a pGEM-T easy vector, the digested PAR1 or PAR2 cDNA fragments were subcloned into the pYesTrp2 vector. The primer set used for cloning is listed in Table S2. All the constructs were sequence verified. The construct pairs were transformed into the yeast, Saccharomyces cerevisiae strain EGY48, and cultured at 30 °C. Successful yeast transformants were cultured in liquid SD minimal medium (-Ura/-His/-Trp) at 230 rpm/30 °C for 24 h so that it reached a final concentration equivalent to OD600, 1.2 to 1.5. Then, 1 mL of yeast strain was centrifuged and resuspended in 0.3 mL of distilled water. Ten microliters of the suspension cells were plated on SD/Gal/Raf (-Ura/-His/-Trp/-Leu) induction medium (MP Biomedicals, Santa Ana, CA, USA). Y2H images were taken 3 d after incubation at 30 °C.
4.6. Promoter Activity Analysis
A 1.2 kb DNA fragment containing the 5′ upstream region of PRE6 was amplified with primers (5′-GGATCCTGCCATTTTAAACCCCACTT-3′ and 5′-CGATCGGTGTTGAAGAGGGAGGAGCA-3′). The amplified fragment was cloned into a pGEM-T Easy vector, after verification of no sequence error, then subcloned into the pCAMBIA1381 vector. The resulting recombinant plasmid was transformed into the Agrobacterium tumefaciens strain GV3101, and introduced into wild type plants using the floral dipping method. Histochemical analysis of GUS activity in Arabidopsis tissues was done as described by Jefferson et al. (1987). The stained tissues were post-fixed in FAA (4% formaldehyde (v/v), 5% acetic acid (v/v), and 50% ethanol (v/v)) and cleared in 70% ethanol before being photographed. Whole-mount tissue samples were viewed with Axioplan microscopes (Carl Zeiss). Digital images were captured using a Spot II camera (Diagnostic Instruments Inc, Starling Height, MI, USA).
5. Conclusions
In summary, our study with loss-of-function pre mutants not only confirmed the regulatory function of PREs in cell elongation during vegetative development but also revealed the role of PREs in cell elongation of anther filament, which is critical for successful pollination. Taking together with the results from gain-of-function studies, we suggest that PREs may constitute versatile growth-regulatory units in various developmental contexts. As exemplified in the previous report that altered expression of a PRE ortholog, Style2.1, is associated with the evolution of self-pollination in cultivated tomato species [], we propose that manipulation of PRE expression using tissue-specific promoters is a promising way to manipulate self-pollination behavior of flowering plants, an agronomically important trait.
Supplementary Materials
Supplementary Materials can be found at http://www.mdpi.com/1422-0067/20/4/869/s1.
Author Contributions
Conceptualization, K.S., M.-S.S., and S.L.; Methodology, K.S., S.K.P. and S.L.; Investigation, K.S., I.L., E.K. and S.L.; Writing–Original Draft, S.L.; Writing–Review & Editing, K.S., I.L., E.K., S.K.P., M.-S.S. and S.L.; Funding Acquisition, M.-S.S., S.K.P., and S.L.; Supervision, S.L. and M.-S.S.
Funding
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2015R1D1A01058039 and 2018R1D1A1B07049914 for M.S.S.), and by the Rural Development Administration [Next-Generation Biogreen21 program (Agricultural Biotechnology Research Center No. PJ01369001) to S.K.P.], and by the Ministry of Science, ICT & Future Planning (2015R1C1A2A01052950 for S.L.).
Acknowledgments
We thank ABRC and NASC for providing the Arabidopsis mutant lines. We are thankful to Jason Reed for providing mutant seeds of myb21 and arf6arf8. We thank Christopher Hicks for editorial suggestions for writing manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| PRE | PACLOBUTRAZOL RESISTANCE |
| WT | Wild type |
| GA | Gibberellin |
| BR | Brassinosteroid |
| JA | Jasmonic acid |
| ARF | AUXIN RESPONSE FACTOR |
| qRT-PCR | Quantitative real-time PCR |
| HLH | Helix loop helix |
References
- Karron, J.D.; Jackson, R.T.; Thumser, N.N.; Schlicht, S.L. Outcrossing rates of individual Mimulus ringens genets are correlated with anther–stigma separation. Heredity 1997, 79, 365. [Google Scholar] [CrossRef]
- Motten, A.F.; Stone, J.L. Heritability of stigma position and the effect of stigma-anther separation on outcrossing in a predominantly self-fertilizing weed, Datura stramonium (Solanaceae). Am. J. Bot. 2000, 87, 339–347. [Google Scholar] [CrossRef]
- Chen, K.Y.; Cong, B.; Wing, R.; Vrebalov, J.; Tanksley, S.D. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 2007, 318, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Cecchetti, V.; Altamura, M.M.; Falasca, G.; Costantino, P.; Cardarelli, M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 2008, 20, 1760–1774. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Gao, H.; Liu, B.; Qi, T.; Tong, J.; Xiao, L.; Xie, D.; Song, S. Arabidopsis MYB24 Regulates Jasmonate-Mediated Stamen Development. Front Plant Sci. 2017, 8, 1525. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Gupta, A.; Singh, D.; Khurana, J.P.; Laxmi, A. Arabidopsis RSS1 Mediates Cross-Talk Between Glucose and Light Signaling During Hypocotyl Elongation Growth. Sci. Rep. 2017, 7, 16101. [Google Scholar] [CrossRef]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Koornneef, M.; van der Veen, J.H. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor. Appl. Genet. 1980, 58, 257–263. [Google Scholar] [CrossRef]
- Mandaokar, A.; Thines, B.; Shin, B.; Lange, B.M.; Choi, G.; Koo, Y.J.; Yoo, Y.J.; Choi, Y.D.; Choi, G.; Browse, J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006, 46, 984–1008. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [PubMed]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Zhu, W.; Li, L.; Zhang, S.; Yin, Y.; Ma, H.; Wang, X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 2010, 107, 6100–6105. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Song, S.; Xiao, L.; Soo, H.M.; Cheng, Z.; Xie, D.; Peng, J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009, 5, e1000440. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.H.; Ellis, C.M.; Ploense, S.E.; Wu, M.F.; Yadav, V.; Tholl, D.; Chetelat, A.; Haupt, I.; Kennerley, B.J.; Hodgens, C.; et al. A regulatory network for coordinated flower maturation. PLoS Genet. 2012, 8, e1002506. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Lee, I. KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 2006, 61, 283–296. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef]
- Mara, C.D.; Huang, T.; Irish, V.F. The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 2010, 22, 690–702. [Google Scholar] [CrossRef]
- Schlereth, A.; Moller, B.; Liu, W.; Kientz, M.; Flipse, J.; Rademacher, E.H.; Schmid, M.; Jurgens, G.; Weijers, D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 2010, 464, 913–916. [Google Scholar] [CrossRef]
- Wang, H.; Zhu, Y.; Fujioka, S.; Asami, T.; Li, J.; Li, J. Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 2009, 21, 3781–3791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.; Zhu, J.Y.; Wang, H.; Zhang, Z.; Wang, W.; Sun, Y.; Zhao, J.; Sun, X.; et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Greenham, K.; Castillejo, C.; Sartor, R.; Bialy, A.; Sun, T.P.; Estelle, M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS ONE 2012, 7, e36210. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 2012, 14, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Fujiwara, S.; Mitsuda, N.; Ohme-Takagi, M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 2012, 24, 4483–4497. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal Behav. 2013, 8, e23448. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; van den Berg, W.; Lokerse, A.; Liao, C.Y.; van Mourik, H.; Moller, B.; Peris, C.L.; Weijers, D. A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant. Physiol. 2011, 156, 1292–1299. [Google Scholar] [CrossRef]
- Hao, Y.; Oh, E.; Choi, G.; Liang, Z.; Wang, Z.Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant. 2012, 5, 688–697. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W.; et al. 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef]
- Nagpal, P.; Ellis, C.M.; Weber, H.; Ploense, S.E.; Barkawi, L.S.; Guilfoyle, T.J.; Hagen, G.; Alonso, J.M.; Cohen, J.D.; Farmer, E.E.; et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 2005, 132, 4107–4118. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Bai, M.Y.; Arenhart, R.A.; Sun, Y.; Wang, Z.Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Wang, Y.; Zhang, N.; Jia, Q.; Wang, X.; Hou, C.; Chen, J.G.; Wang, S. Involvement of PACLOBUTRAZOL RESISTANCE6/KIDARI, an Atypical bHLH Transcription Factor, in Auxin Responses in Arabidopsis. Front. Plant. Sci. 2017, 8, 1813. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant. J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Mitina, I.; Quach, H.L.; Theologis, A. AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant. J. 2005, 43, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Tashiro, S.; Tian, C.E.; Watahiki, M.K.; Yamamoto, K.T. Changes in growth kinetics of stamen filaments cause inefficient pollination in massugu2, an auxin insensitive, dominant mutant of Arabidopsis thaliana. Physiol. Plant. 2009, 137, 175–187. [Google Scholar] [CrossRef]
- Zhiponova, M.K.; Morohashi, K.; Vanhoutte, I.; Machemer-Noonan, K.; Revalska, M.; Van Montagu, M.; Grotewold, E.; Russinova, E. Helix-loop-helix/basic helix-loop-helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proc. Natl. Acad. Sci. USA 2014, 111, 2824–2829. [Google Scholar] [CrossRef]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant. Cell 1990, 2, 755–767. [Google Scholar] [CrossRef]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant. Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant. J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).