The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tagtm Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process
Abstract
:1. Introduction
2. Results
2.1. Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tagTM Agarose
2.2. Antibody Preparation and Evaluation of AGPase Small Subunit Bt2
2.3. Co-Immunoprecipitation of Stromal Protein and Expression Analysis of Bt2 in Maize Endosperm
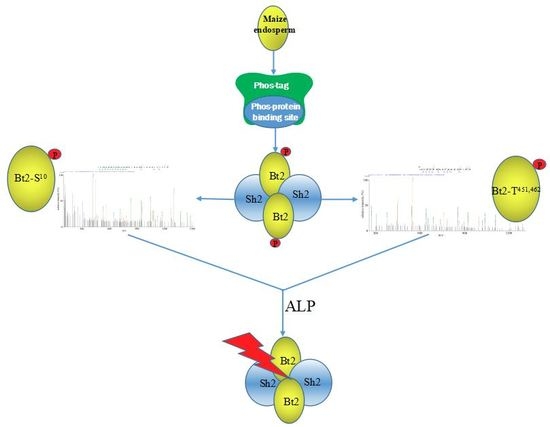
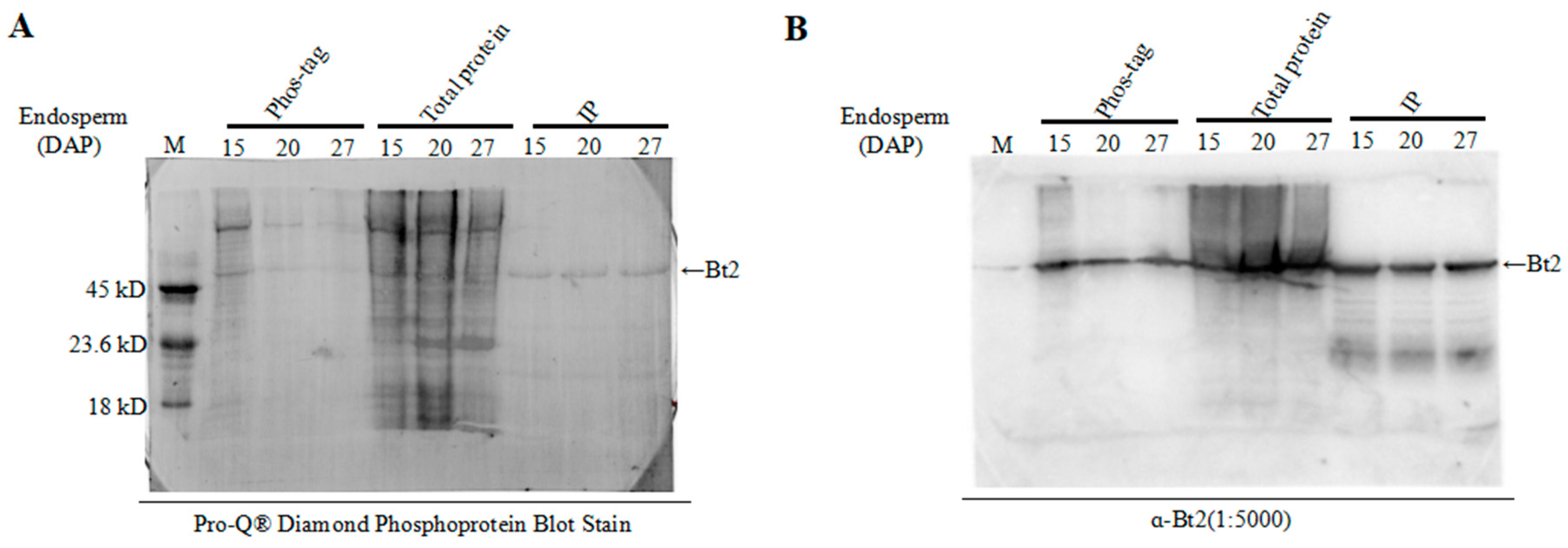
2.4. Enrichment of Bt2 Phosphorylation Protein by Phos-tagTM
2.5. Detection of Bt2 Phosphorylation Protein by Diamond Q Staining Technology
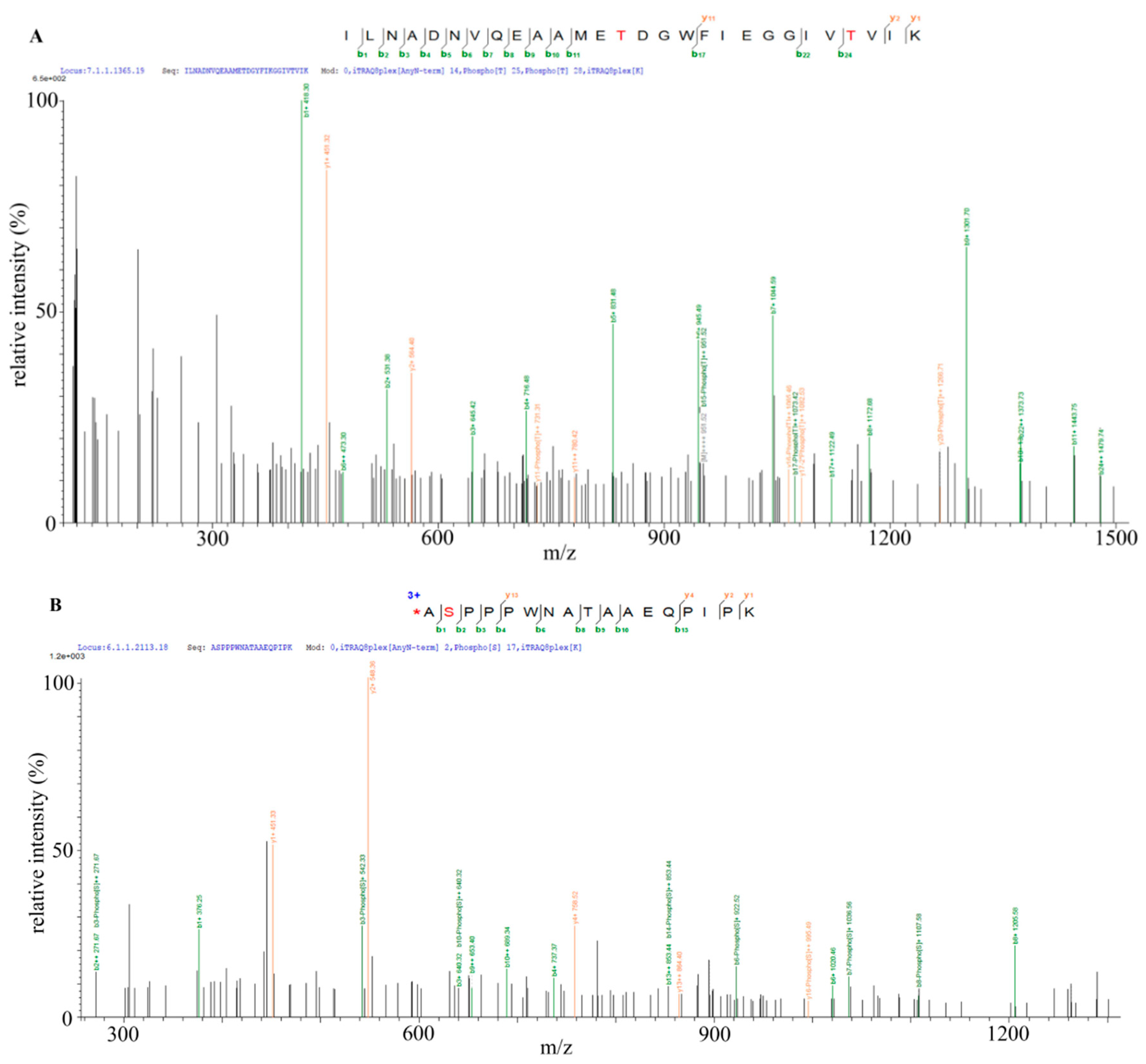
2.6. Identification of Bt2 Phosphorylation Sites by iTRAQ
2.7. Enzyme Characteristics of AGPase Phosphorylation and a Potential Regulatory Model
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. GST-Gene Fusion System Protein Expression and Purification
4.3. Rabbit Breeding, Anti-serum Preparation, and Antibody Purification
4.4. Plant Protein Extraction and Protein Determination
4.5. SDS-PAGE and Immunoblotting
4.6. Mass Spectrometry and Data Processing
4.7. Immunoprecipitation (IP) and Co-immunoprecipitation (Co-IP)
4.8. Pro-Q Diamond Phosphoprotein Staining
4.9. iTRAQTM Labeling and Mass Spectrometry Analysis
4.10. Zymograms of Native PAGE
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| Co-IP | Co-immunoprecipitation |
| DAP | Days After Pollination |
| GBSS | Granule bound starch synthase |
| IP | immunoprecipitation |
| SBEI | Starch Branch Enzyme I |
| SBEIIa | Starch Branch Enzyme IIa |
| SP | Starch Phosphorylase |
| SSI | Starch synthase I |
| SSII | Starch synthase II |
References
- Chen, J.; Yi, Q.; Cao, Y.; Wei, B.; Zheng, L.; Xiao, Q.; Xie, Y.; Gu, Y.; Li, Y.; Huang, H.; et al. ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 2016, 67, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Hannah, L.C.; James, M. The complexities of starch biosynthesis in cereal endosperms. Curr. Opin. Biotechnol. 2008, 19, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Denyer, K.; Martin, C. The Synthesis of the Starch Granule. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Georgelis, N.; Braun, E.L.; Shaw, J.R.; Hannah, L.C. The two AGPase subunits evolve at different rates in angiosperms, yet they are equally sensitive to activity-altering amino acid changes when expressed in bacteria. Plant Cell 2007, 19, 1458–1472. [Google Scholar] [CrossRef] [PubMed]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Nerlich, A.; Faix, B.; Hummer, C.; Fox, S.; Trafford, K.; Weber, H.; Weschke, W.; Geigenberger, P. Subcellular analysis of starch metabolism in developing barley seeds using a non-aqueous fractionation method. J. Exp. Bot. 2012, 63, 2071–2087. [Google Scholar] [CrossRef] [PubMed]
- Hannah, L.C.; Tuschall, D.M.; Mans, R.J. Multiple forms of maize endosperm adp-glucose pyrophosphorylase and their control by shrunken-2 and brittle-2. Genetics 1980, 95, 961–970. [Google Scholar] [PubMed]
- Hannah, L.C.; Shaw, J.R.; Giroux, M.J.; Reyss, A.; Prioul, J.L.; Bae, J.M.; Lee, J.Y. Maize genes encoding the small subunit of ADP-glucose pyrophosphorylase. Plant Physiol. 2001, 127, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Greene, T.W.; Hannah, L.C. Enhanced stability of maize endosperm ADP-glucose pyrophosphorylase is gained through mutants that alter subunit interactions. Proc. Natl. Acad. Sci. USA 1998, 95, 13342–13347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linebarger, C.R.; Boehlein, S.K.; Sewell, A.K.; Shaw, J.; Hannah, L.C. Heat stability of maize endosperm ADP-glucose pyrophosphorylase is enhanced by insertion of a cysteine in the N terminus of the small subunit. Plant Physiol. 2005, 139, 1625–1634. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Shaw, J.R.; Stewart, J.D.; Hannah, L.C. Studies of the kinetic mechanism of maize endosperm ADP-glucose pyrophosphorylase uncovered complex regulatory properties. Plant Physiol. 2010, 152, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Shaw, J.R.; Georgelis, N.; Hannah, L.C. Enhanced heat stability and kinetic parameters of maize endosperm ADPglucose pyrophosphorylase by alteration of phylogenetically identified amino acids. Arch. Biochem. Biophys. 2014, 543, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Burger, B.T.; Cross, J.M.; Shaw, J.R.; Caren, J.R.; Greene, T.W.; Okita, T.W.; Hannah, L.C. Relative turnover numbers of maize endosperm and potato tuber ADP-glucose pyrophosphorylases in the absence and presence of 3-phosphoglyceric acid. Planta 2003, 217, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.M.; Clancy, M.; Shaw, J.R.; Boehlein, S.K.; Greene, T.W.; Schmidt, R.R.; Okita, T.W.; Hannah, L.C. A polymorphic motif in the small subunit of ADP-glucose pyrophosphorylase modulates interactions between the small and large subunits. Plant J. 2005, 41, 501–511. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Shaw, J.R.; Stewart, J.D.; Hannah, L.C. Heat stability and allosteric properties of the maize endosperm ADP-glucose pyrophosphorylase are intimately intertwined. Plant Physiol. 2008, 146, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Sowokinos, J.R. Pyrophosphorylases in Solanum tuberosum: II. Catalytic properties and regulation of ADP-glucose and UDP-glucose pyrophosphorylase activities in potatoes. Plant Physiol. 1981, 68, 924–929. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.M.; Clancy, M.; Shaw, J.R.; Greene, T.W.; Schmidt, R.R.; Okita, T.W.; Hannah, L.C. Both subunits of ADP-glucose pyrophosphorylase are regulatory. Plant Physiol. 2004, 135, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Sowokinos, J.R.; Preiss, J. Pyrophosphorylases in Solanum tuberosum: III. Purification, Physical, and Catalytic Properties of Adpglucose Pyrophosphorylase in Potatoes. Plant Physiol. 1982, 69, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Shaw, J.R.; McCarty, D.R.; Hwang, S.K.; Stewart, J.D.; Hannah, L.C. The potato tuber, maize endosperm and a chimeric maize-potato ADP-glucose pyrophosphorylase exhibit fundamental differences in Pi inhibition. Arch. Biochem. Biophys. 2013, 537, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Hannah, L.C.; Futch, B.; Bing, J.; Shaw, J.R.; Boehlein, S.; Stewart, J.D.; Beiriger, R.; Georgelis, N.; Greene, T. A shrunken-2 transgene increases maize yield by acting in maternal tissues to increase the frequency of seed development. Plant Cell 2012, 24, 2352–2363. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhang, S.; Zhao, Y.; Li, B.; Zhang, J. Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 2011, 233, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Burrell, M.M.; ap Rees, T. Starch metabolism in tubers of transgenic potato (Solanum tuberosum) with increased ADPglucose pyrophosphorylase. Biochem. J. 1996, 320, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Tiessen, A.; Hendriks, J.H.; Stitt, M.; Branscheid, A.; Gibon, Y.; Farre, E.M.; Geigenberger, P. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: A novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 2002, 14, 2191–2213. [Google Scholar] [CrossRef] [PubMed]
- Hendriks, J.H.; Kolbe, A.; Gibon, Y.; Stitt, M.; Geigenberger, P. ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 2003, 133, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gong, F.; Cao, D.; Hu, X.; Wang, W. Advances in crop proteomics: PTMs of proteins under abiotic stress. Proteomics 2016, 16, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Burnell, J.N.; Hatch, M.D. Activation and inactivation of an enzyme catalyzed by a single, bifunctional protein: A new example and why. Arch. Biochem. Biophys. 1986, 245, 297–304. [Google Scholar] [CrossRef]
- Pesaresi, P.; Pribil, M.; Wunder, T.; Leister, D. Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: The roles of STN7, STN8 and TAP38. Biochim. Biophys. Acta 2011, 1807, 887–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tetlow, I.J.; Wait, R.; Lu, Z.; Akkasaeng, R.; Bowsher, C.G.; Esposito, S.; Kosar-Hashemi, B.; Morell, M.K.; Emes, M.J. Protein phosphorylation in amyloplasts regulates starch branching enzyme activity and protein-protein interactions. Plant Cell 2004, 16, 694–708. [Google Scholar] [CrossRef] [PubMed]
- Tetlow, I.J.; Morell, M.K.; Emes, M.J. Recent developments in understanding the regulation of starch metabolism in higher plants. J. Exp. Bot. 2004, 55, 2131–2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Makhmoudova, A.; Lee, E.A.; Wait, R.; Emes, M.J.; Tetlow, I.J. The amylose extender mutant of maize conditions novel protein-protein interactions between starch biosynthetic enzymes in amyloplasts. J. Exp. Bot. 2009, 60, 4423–4440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Ahmed, Z.; Lee, E.A.; Donner, E.; Liu, Q.; Ahmed, R.; Morell, M.K.; Emes, M.J.; Tetlow, I.J. Allelic variants of the amylose extender mutation of maize demonstrate phenotypic variation in starch structure resulting from modified protein-protein interactions. J. Exp. Bot. 2012, 63, 1167–1183. [Google Scholar] [CrossRef] [PubMed]
- Makhmoudova, A.; Williams, D.; Brewer, D.; Massey, S.; Patterson, J.; Silva, A.; Vassall, K.A.; Liu, F.; Subedi, S.; Harauz, G.; et al. Identification of multiple phosphorylation sites on maize endosperm starch branching enzyme IIb, a key enzyme in amylopectin biosynthesis. J. Biol. Chem. 2014, 289, 9233–9246. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Shen, Z.; Sartor, R.; Wu, K.J.; Osborn, J.; Smith, L.G.; Briggs, S.P. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc. Natl. Acad. Sci. USA 2013, 110, E4808–E4817. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Hennen-Bierwagen, T.A.; Myers, A.M. Functions of multiple genes encoding ADP-glucose pyrophosphorylase subunits in maize endosperm, embryo, and leaf. Plant Physiol. 2014, 164, 596–611. [Google Scholar] [CrossRef] [PubMed]
- Goodman, T.; Schulenberg, B.; Steinberg, T.H.; Patton, W.F. Detection of phosphoproteins on electroblot membranes using a small-molecule organic fluorophore. Electrophoresis 2004, 25, 2533–2538. [Google Scholar] [CrossRef] [PubMed]
- Schulenberg, B.; Goodman, T.N.; Aggeler, R.; Capaldi, R.A.; Patton, W.F. Characterization of dynamic and steady-state protein phosphorylation using a fluorescent phosphoprotein gel stain and mass spectrometry. Electrophoresis 2004, 25, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Dinges, J.R.; Colleoni, C.; Myers, A.M.; James, M.G. Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 2001, 125, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Wakuta, S.; Shibata, Y.; Yoshizaki, Y.; Saburi, W.; Hamada, S.; Ito, H.; Hwang, S.K.; Okita, T.W.; Matsui, H. Modulation of allosteric regulation by E38K and G101N mutations in the potato tuber ADP-glucose pyrophosphorylase. Biosci. Biotechnol. Biochem. 2013, 77, 1854–1859. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Kubota, Y.; Takekawa, M.; Koike, T. A Phos-tag SDS-PAGE method that effectively uses phosphoproteomic data for profiling the phosphorylation dynamics of MEK1. Proteomics 2016, 16, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, E.; Kinoshita-Kikuta, E.; Koike, T. Phosphate-affinity polyacrylamide gel electrophoresis for SNP genotyping. Meth. Mol. Biol. 2009, 578, 183–192. [Google Scholar]
- Kinoshita-Kikuta, E.; Kinoshita, E.; Yamada, A.; Endo, M.; Koike, T. Enrichment of phosphorylated proteins from cell lysate using a novel phosphate-affinity chromatography at physiological pH. Proteomics 2006, 6, 5088–5095. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, P.; Hertle, A.; Pribil, M.; Schneider, A.; Kleine, T.; Leister, D. Optimizing photosynthesis under fluctuating light: The role of the Arabidopsis STN7 kinase. Plant Signal. Behav. 2010, 5, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Pribil, M.; Pesaresi, P.; Hertle, A.; Barbato, R.; Leister, D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 2010, 8, e1000288. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Batra, R.; Saripalli, G.; Mohan, A.; Gupta, S.; Gill, K.S.; Varadwaj, P.K.; Balyan, H.S.; Gupta, P.K. Comparative Analysis of AGPase Genes and Encoded Proteins in Eight Monocots and Three Dicots with Emphasis on Wheat. Front. Plant Sci. 2017, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Ballicora, M.A.; Preiss, J.; Geiger, J.H. Crystal structure of potato tuber ADP-glucose pyrophosphorylase. EMBO J. 2005, 24, 694–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comino, N.; Cifuente, J.O.; Marina, A.; Orrantia, A.; Eguskiza, A.; Guerin, M.E. Mechanistic insights into the allosteric regulation of bacterial ADP-glucose pyrophosphorylases. J. Biol. Chem. 2017, 292, 6255–6268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Wu, M.; Pei, W.; Li, H.; Li, X.; Zhang, J.; Yu, J.; Yu, S. Quantitative phosphoproteomic profiling of fiber differentiation and initiation in a fiberless mutant of cotton. BMC Genom. 2014, 15, 466. [Google Scholar] [CrossRef] [PubMed]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.; Lv, Y.; Shen, L.; Wang, Y.; Qing, Y.; Wu, N.; Li, Y.; Huang, H.; Zhang, N.; Liu, Y.; et al. The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tagtm Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. Int. J. Mol. Sci. 2019, 20, 986. https://doi.org/10.3390/ijms20040986
Yu G, Lv Y, Shen L, Wang Y, Qing Y, Wu N, Li Y, Huang H, Zhang N, Liu Y, et al. The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tagtm Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. International Journal of Molecular Sciences. 2019; 20(4):986. https://doi.org/10.3390/ijms20040986
Chicago/Turabian StyleYu, Guowu, Yanan Lv, Leiyang Shen, Yongbin Wang, Yun Qing, Nan Wu, Yangping Li, Huanhuan Huang, Na Zhang, Yinghong Liu, and et al. 2019. "The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tagtm Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process" International Journal of Molecular Sciences 20, no. 4: 986. https://doi.org/10.3390/ijms20040986
APA StyleYu, G., Lv, Y., Shen, L., Wang, Y., Qing, Y., Wu, N., Li, Y., Huang, H., Zhang, N., Liu, Y., Hu, Y., Liu, H., Zhang, J., & Huang, Y. (2019). The Proteomic Analysis of Maize Endosperm Protein Enriched by Phos-tagtm Reveals the Phosphorylation of Brittle-2 Subunit of ADP-Glc Pyrophosphorylase in Starch Biosynthesis Process. International Journal of Molecular Sciences, 20(4), 986. https://doi.org/10.3390/ijms20040986