Interactions between WUSCHEL- and CYC2-like Transcription Factors in Regulating the Development of Reproductive Organs in Chrysanthemum morifolium
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of CmWUS
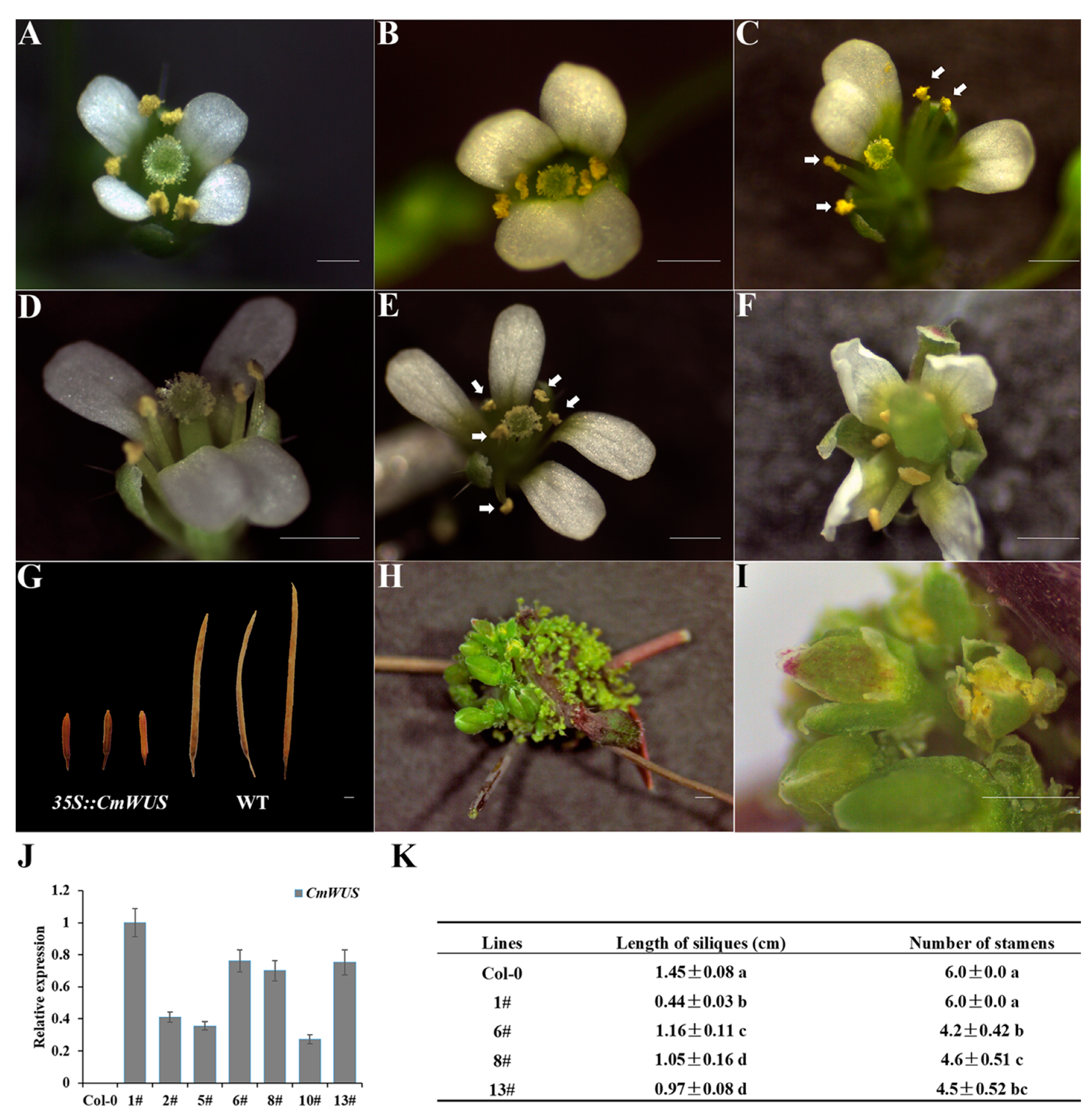
2.2. Overexpression of CmWUS in A. thaliana Inhibits the Development of Reproductive Organs and Affects Flower Symmetry
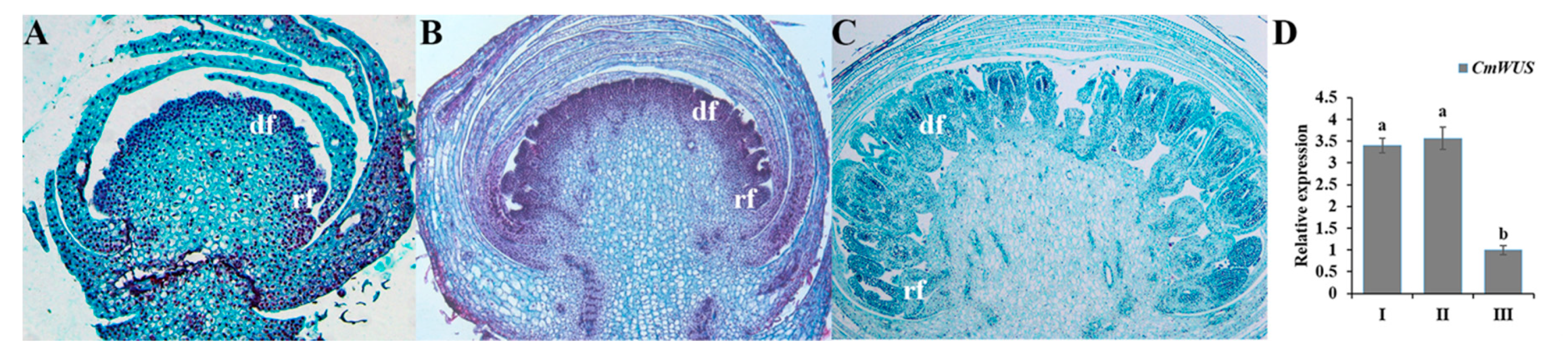
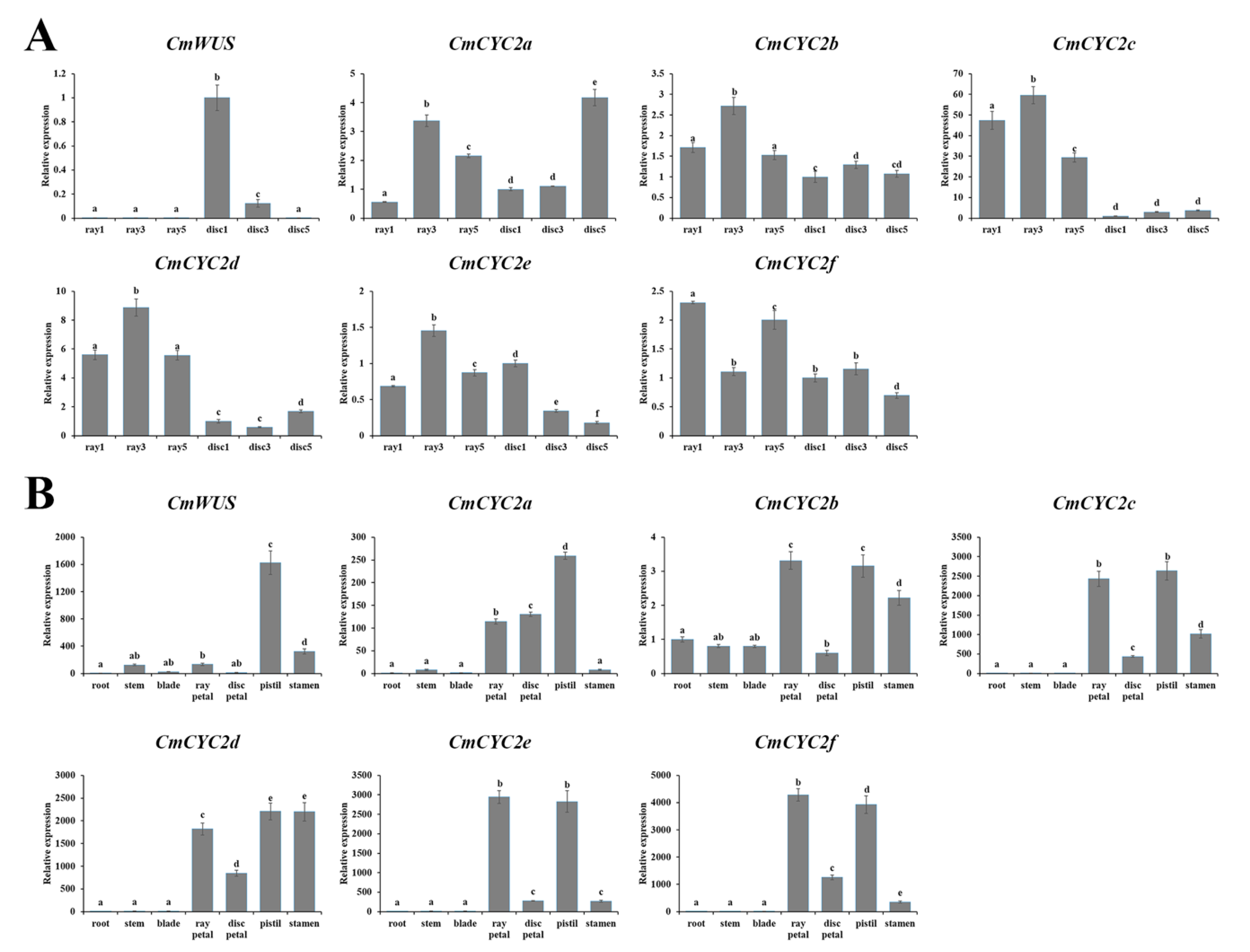
2.3. High Expression of CmWUS and CmCYC2 in the Reproductive Organs of C. morifolium
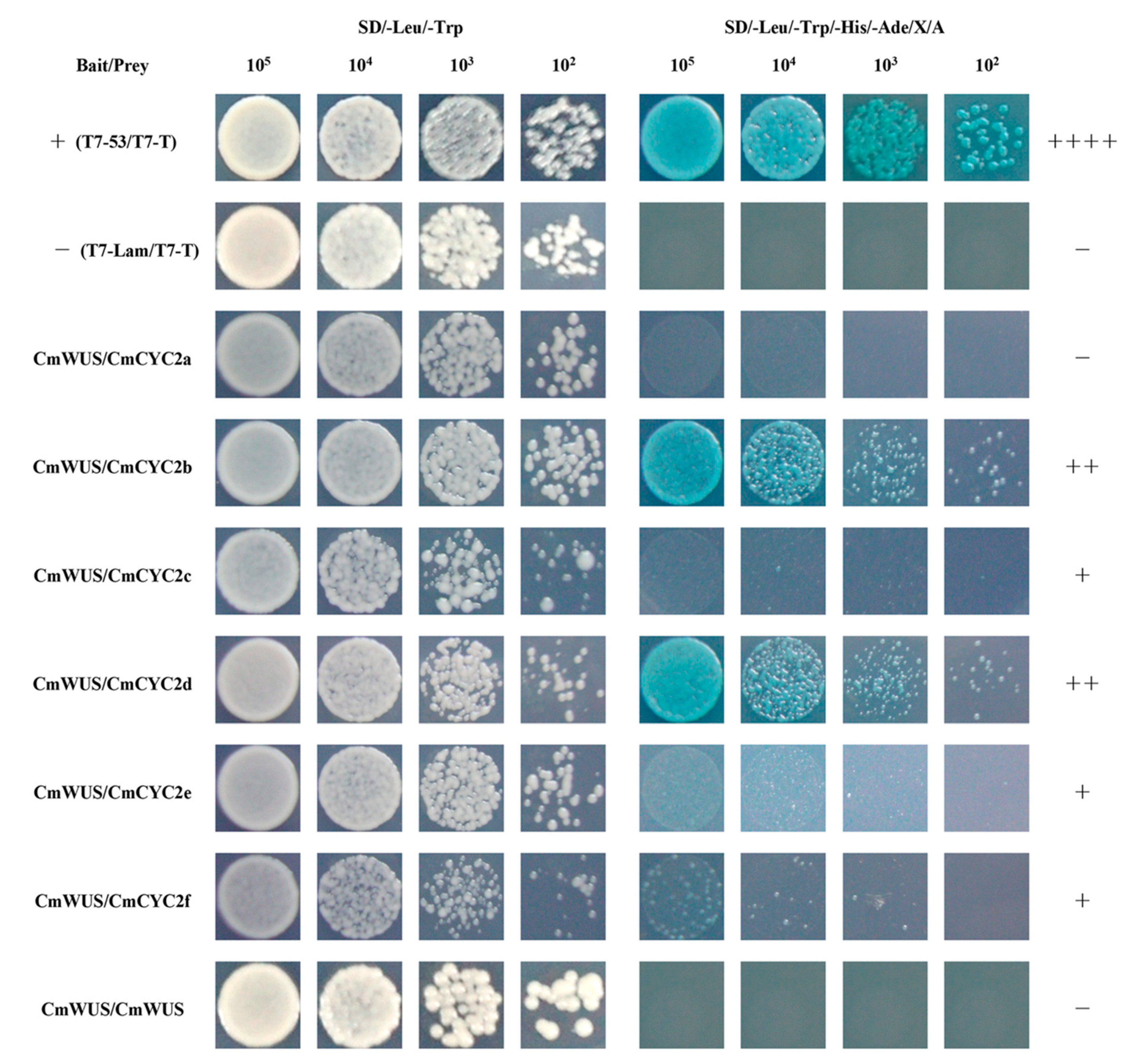
2.4. Protein-Protein Interactions between CmWUS and CmCYC2
3. Discussion
3.1. Ectopic Expression of CmWUS in A. thaliana Indicates Possible Conserved Functions in Floral Meristems
3.2. Proposed Interaction between CmWUS and CmCYC2 in Regulating Reproductive Organ Development
3.3. WUS Can Be a Bridge to Connect MADS-box and ECE (CYC/TB1)
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Gene Cloning
4.3. Bioinformatics Analysis
4.4. Overexpression of CmWUS in A. thaliana
4.5. Microscope Observations
4.6. Gene Expression Analysis in C. morifolium
4.7. Subcellular Localization
4.8. Y2H Assay
4.9. BiFC Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gillies, A.C.M.; Cubas, P.; Coen, E.S.; Abbott, R.J. Making rays in the Asteraceae: Genetics and evolution of radiate versus discoid flower heads. In Developmental Genetics and Plant Evolution; Quentin, C.B., Cronk, R.M.B., Julie, A.H., Eds.; Taylor & Francis: London, UK, 2002; Volume 65, pp. 233–246. [Google Scholar]
- Bertin, R.I.; Kerwin, M.A. Floral sex ratios and gynomonoecy in Aster (Asteraceae). Am. J. Bot. 1998, 85, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Bertin, R.I.; Connors, D.B.; Kleinman, H.M. Differential herbivory on disk and ray flowers of gynomonoecious asters and goldenrods (Asteraceae). Biol. J. Linn. Soc. 2010, 101, 544–552. [Google Scholar] [CrossRef]
- Sun, M.; Ganders, F.R. Outcrossing rates and allozyme variation in rayed and rayless morphs of Bidens pilosa. Heredity 1990, 64, 139–143. [Google Scholar] [CrossRef]
- Andersson, S. Pollinator and nonpollinator selection on ray morphology in Leucanthemum vulgare (oxeye daisy, Asteraceae). Am. J. Bot. 2008, 95, 1072–1078. [Google Scholar] [CrossRef]
- Fambrini, M.; Pugliesi, C. CYCLOIDEA 2 clade genes: Key players in the control of floral symmetry, inflorescence architecture, and reproductive organ development. Plant Mol. Biol. Rep. 2016, 35, 20–36. [Google Scholar] [CrossRef]
- Kalisz, S.; Ree, R.H.; Sargent, R.D. Linking floral symmetry genes to breeding system evolution. Trends Plant Sci. 2006, 11, 568–573. [Google Scholar] [CrossRef]
- Hileman, L.C.; Cubas, P. An expanded evolutionary role for flower symmetry genes. J. Biol. 2009, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef]
- Martin-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar] [CrossRef]
- Hileman, L.C. Bilateral flower symmetry—How, when and why? Curr. Opin. Plant Biol. 2014, 17, 146–152. [Google Scholar] [CrossRef]
- Broholm, S.K.; Teeri, T.H.; Elomaa, P. Molecular control of inflorescence development in Asteraceae. In Advances in Botanical Research; Fornara, F., Ed.; Academic Press: Oxford, UK, 2014; Volume 72, pp. 297–333. [Google Scholar]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Copsey, L.; Vincent, C.; Clark, J.; Coen, E. Control of organ asymmetry in flowers of Antirrhinum. Cell 1999, 99, 367–376. [Google Scholar] [CrossRef]
- Gaudin, V.; Lunness, P.A.; Fobert, P.R.; Towers, M.; Rioukhamlichi, C.; Murray, J.A.H.; Coen, E.; Doonan, J.H. The Expression of D-Cyclin Genes Defines Distinct Developmental Zones in Snapdragon Apical Meristems and Is Locally Regulated by the Cycloidea Gene. Plant Physiol. 2000, 122, 1137–1148. [Google Scholar] [CrossRef]
- Song, C.F.; Lin, Q.B.; Liang, R.H.; Wang, Y.Z. Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae). BMC Evol. Biol. 2009, 9, 244. [Google Scholar] [CrossRef]
- Preston, J.C.; Kost, M.A.; Hileman, L.C. Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol. 2009, 182, 751–762. [Google Scholar] [CrossRef]
- Zhao, Y.; Pfannebecker, K.; Dommes, A.B.; Hidalgo, O.; Becker, A.; Elomaa, P. Evolutionary diversification of CYC/TB1-like TCP homologs and their recruitment for the control of branching and floral morphology in Papaveraceae (basal eudicots). New Phytol. 2018, 220, 317–331. [Google Scholar] [CrossRef]
- Broholm, S.K.; Tahtiharju, S.; Laitinen, R.A.; Albert, V.A.; Teeri, T.H.; Elomaa, P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 9117–9122. [Google Scholar] [CrossRef]
- Juntheikki-Palovaara, I.; Tahtiharju, S.; Lan, T.; Broholm, S.K.; Rijpkema, A.S.; Ruonala, R.; Kale, L.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. 2014, 79, 783–796. [Google Scholar] [CrossRef]
- Elomaa, P.; Zhao, Y.; Zhang, T. Flower heads in Asteraceae—Recruitment of conserved developmental regulators to control the flower-like inflorescence architecture. Hortic. Res. 2018, 5, 36. [Google Scholar] [CrossRef]
- Tahtiharju, S.; Rijpkema, A.S.; Vetterli, A.; Albert, V.A.; Teeri, T.H.; Elomaa, P. Evolution and diversification of the CYC/TB1 gene family in Asteraceae—A comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol. Biol. Evol. 2012, 29, 1155–1166. [Google Scholar] [CrossRef]
- Bello, M.A.; Cubas, P.; Alvarez, I.; Sanjuanbenito, G.; Fuertes-Aguilar, J. Evolution and expression patterns of CYC/TB1 genes in Anacyclus: Phylogenetic insights for floral symmetry genes in Asteraceae. Front. Plant Sci. 2017, 8, 589. [Google Scholar] [CrossRef]
- Mizzotti, C.; Fambrini, M.; Caporali, E.; Masiero, S.; Pugliesi, C. A CYCLOIDEA-like gene mutation in sunflower determines an unusual floret type able to produce filled achenes at the periphery of the pseudanthium. Botany 2015, 93, 171–181. [Google Scholar] [CrossRef]
- Fambrini, M.; Salvini, M.; Pugliesi, C. A transposon-mediate inactivation of a CYCLOIDEA-like gene originates polysymmetric and androgynous ray flowers in Helianthus Annuus. Genetics 2011, 139, 1521–1529. [Google Scholar] [CrossRef]
- Chapman, M.A.; Tang, S.; Draeger, D.; Nambeesan, S.; Shaffer, H.; Barb, J.G.; Knapp, S.J.; Burke, J.M. Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PLoS Genet. 2012, 8, e1002628. [Google Scholar] [CrossRef]
- Fambrini, M.; Basile, A.; Salvini, M.; Pugliesi, C. Excisions of a defective transposable CACTA element (Tetu1) generate new alleles of a CYCLOIDEA-like gene of Helianthus annuus. Gene 2014, 549, 198–207. [Google Scholar] [CrossRef]
- Liu, H.; Sun, M.; Du, D.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q.; Gao, Y. Whole-transcriptome analysis of differentially expressed genes in the ray florets and disc florets of Chrysanthemum morifolium. BMC Genom. 2016, 17, 398. [Google Scholar] [CrossRef]
- Graaff, E.V.D.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genom. Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A Molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2012, 25, 2025–2030. [Google Scholar] [CrossRef]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef]
- Deyhle, F.; Sarkar, A.K.; Tucker, E.J.; Laux, T. WUSCHEL regulates cell differentiation during anther development. Dev. Biol. 2007, 302, 154–159. [Google Scholar] [CrossRef]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genom. Res. 2002, 12, 47–56. [Google Scholar] [CrossRef]
- Yamada, T.; Sasaki, Y.; Hashimoto, K.; Nakajima, K.; Gasser, C.S. CORONA, PHABULOSA and PHAVOLUTA collaborate with BELL1 to confine WUSCHEL expression to the nucellus in Arabidopsis ovules. Development 2016, 143, 422–426. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar]
- Liu, X.; Ning, K.; Che, G.; Yan, S.; Han, L.; Gu, R.; Li, Z.; Weng, Y.; Zhang, X. CsSPL functions as an adaptor between HD-ZIP III and CsWUS transcription factors regulating anther and ovule development in Cucumis sativus (cucumber). Plant J. 2018, 94, 535–547. [Google Scholar] [CrossRef]
- Huang, D.; Li, X.; Sun, M.; Zhang, T.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Identification and Characterization of CYC-Like Genes in Regulation of Ray Floret Development in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 1633. [Google Scholar] [CrossRef]
- Ikeda, M.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 2009, 21, 3493–3505. [Google Scholar] [CrossRef]
- Gallois, J.L.; Nora, F.R.; Mizukami, Y.; Sablowski, R. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 2004, 18, 375–380. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Wang, X.M.; Li, J.; Li, J.H.; Wu, J.S.; Walker, J.C.; Xu, Z.H.; Chong, K. Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol. Biol. 2005, 57, 773–784. [Google Scholar] [CrossRef]
- Rodriguez, K.; Perales, M.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. DNA-dependent homodimerization, sub-cellular partitioning, and protein destabilization control WUSCHEL levels and spatial patterning. Proc. Natl. Acad. Sci. USA 2016, 113, 6307–6315. [Google Scholar] [CrossRef]
- Paponov, I.A.; Teale, W.; Lang, D.; Paponov, M.; Reski, R.; Rensing, S.A.; Palme, K. The evolution of nuclear auxin signalling. BMC Evol. Biol. 2009, 9, 126. [Google Scholar] [CrossRef]
- Lenhard, M.; Bohnert, A.; Jurgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef]
- Honda, E.; Yew, C.L.; Yoshikawa, T.; Sato, Y.; Hibara, K.I.; Itoh, J.I. LEAF LATERAL SYMMETRY1, a member of the WUSCHEL-RELATED HOMEOBOX3 gene family, regulates lateral organ development differentially from other paralogs, NARROW LEAF2 and NARROW LEAF3 in Rice. Plant Cell Physiol. 2018, 59, 376–391. [Google Scholar] [CrossRef]
- Berti, F.; Fambrini, M.; Turi, M.; Bertini, D.; Pugliesi, C. Mutations of corolla symmetry affect carpel and stamen development in Helianthus annuus. Can. J. Bot. 2005, 83, 1065–1072. [Google Scholar] [CrossRef]
- Preston, J.C.; Hileman, L.C. Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. Evodevo 2012, 3, 6. [Google Scholar] [CrossRef]
- Clark, J.I.; Coen, E.S. The cycloidea gene can respond to a common dorsoventral prepattern in Antirrhinum. Plant J. 2002, 30, 639–648. [Google Scholar] [CrossRef]
- Zhu, B.; Li, H.; Wen, J.; Mysore, K.S.; Wang, X.; Pei, Y.; Niu, L.; Lin, H. Functional specialization of duplicated AGAMOUS homologs in regulating floral organ development of Medicago truncatula. Front. Plant Sci. 2018, 9, 854. [Google Scholar] [CrossRef]
- Ruokolainen, S.; Ng, Y.P.; Albert, V.A.; Elomaa, P.; Teeri, T.H. Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 2011, 107, 1491–1499. [Google Scholar] [CrossRef]
- Liu, X.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.; Pan, Y.; Cao, X.; Goodrich, J.; Chen, X. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef]
- Huang, Z.; Shi, T.; Zheng, B.; Yumul, R.E.; Liu, X.; You, C.; Gao, Z.; Xiao, L.; Chen, X. APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol. 2017, 215, 1197–1209. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Gu, C.; Chen, S.; Liu, Z.; Shan, H.; Luo, H.; Guan, Z.; Chen, F. Reference gene selection for quantitative real-time PCR in Chrysanthemum subjected to biotic and abiotic stress. Mol. Biotechnol. 2011, 49, 192–197. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zhao, K.; Zhou, Y.; Ahmad, S.; Xu, Z.; Li, Y.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. Comprehensive cloning of Prunus mume dormancy associated MADS-Box genes and their response in flower bud development and dormancy. Front. Plant Sci. 2018, 9, 17. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, Z.; Yong, X.; Ahmad, S.; Yang, W.; Cheng, T.; Wang, J.; Zhang, Q. SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biol. 2017, 17, 10. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Sun, M.; Yuan, C.; Han, Y.; Zheng, T.; Cheng, T.; Wang, J.; Zhang, Q. Interactions between WUSCHEL- and CYC2-like Transcription Factors in Regulating the Development of Reproductive Organs in Chrysanthemum morifolium. Int. J. Mol. Sci. 2019, 20, 1276. https://doi.org/10.3390/ijms20061276
Yang Y, Sun M, Yuan C, Han Y, Zheng T, Cheng T, Wang J, Zhang Q. Interactions between WUSCHEL- and CYC2-like Transcription Factors in Regulating the Development of Reproductive Organs in Chrysanthemum morifolium. International Journal of Molecular Sciences. 2019; 20(6):1276. https://doi.org/10.3390/ijms20061276
Chicago/Turabian StyleYang, Yi, Ming Sun, Cunquan Yuan, Yu Han, Tangchun Zheng, Tangren Cheng, Jia Wang, and Qixiang Zhang. 2019. "Interactions between WUSCHEL- and CYC2-like Transcription Factors in Regulating the Development of Reproductive Organs in Chrysanthemum morifolium" International Journal of Molecular Sciences 20, no. 6: 1276. https://doi.org/10.3390/ijms20061276
APA StyleYang, Y., Sun, M., Yuan, C., Han, Y., Zheng, T., Cheng, T., Wang, J., & Zhang, Q. (2019). Interactions between WUSCHEL- and CYC2-like Transcription Factors in Regulating the Development of Reproductive Organs in Chrysanthemum morifolium. International Journal of Molecular Sciences, 20(6), 1276. https://doi.org/10.3390/ijms20061276





