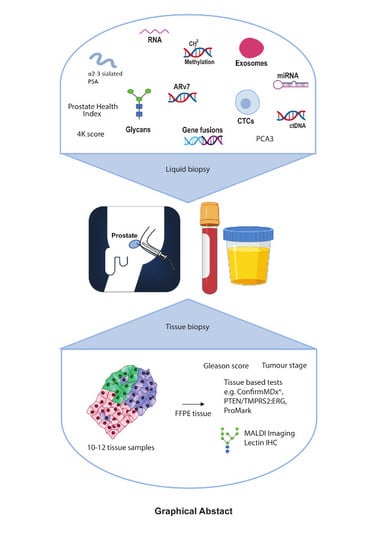
Glycans as Biomarkers in Prostate Cancer
Abstract
:1. Introduction
2. PSA Glycosylation
3. Sialyled Glycans
4. Fucosylation
5. O-GlcNAcylation
6. Branched and Cryptic N-Glycans
7. The F77 Antigen
8. Glycolipids
9. Proteoglycans
10. Galectins
11. Upregulation of Glycosylation Enzymes
12. Exosomes
13. Tissue Imaging of Glycans
14. A Multi-Omic Liquid Biopsy Test
15. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- Attard, G.; Parker, C.; Eeles, R.A.; Schroder, F.; Tomlins, S.A.; Tannock, I.; Drake, C.G.; de Bono, J.S. Prostate cancer. Lancet 2016, 387, 70–82. [Google Scholar] [CrossRef]
- Jamaspishvili, T.; Kral, M.; Khomeriki, I.; Student, V.; Kolar, Z.; Bouchal, J. Urine markers in monitoring for prostate cancer. Prostate Cancer Prostatic Dis. 2010, 13, 12–19. [Google Scholar] [CrossRef]
- Drake, R.R.; White, K.Y.; Fuller, T.W.; Igwe, E.; Clements, M.A.; Nyalwidhe, J.O.; Given, R.W.; Lance, R.S.; Semmes, O.J. Clinical collection and protein properties of expressed prostatic secretions as a source for biomarkers of prostatic disease. J. Proteom. 2009, 72, 907–917. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2018, 20, 71–88. [Google Scholar] [CrossRef]
- Adamczyk, B.; Tharmalingam, T.; Rudd, P.M. Glycans as cancer biomarkers. Biochim. Biophys. Acta 2012, 1820, 1347–1353. [Google Scholar] [CrossRef]
- Reis, C.A.; Osorio, H.; Silva, L.; Gomes, C.; David, L. Alterations in glycosylation as biomarkers for cancer detection. J. Clin. Pathol. 2010, 63, 322–329. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Murphy, K.; Murphy, B.T.; Boyce, S.; Flynn, L.; Gilgunn, S.; O’Rourke, C.J.; Rooney, C.; Stockmann, H.; Walsh, A.L.; Finn, S.; et al. Integrating biomarkers across omic platforms: An approach to improve stratification of patients with indolent and aggressive prostate cancer. Mol. Oncol. 2018, 12, 1513–1525. [Google Scholar] [CrossRef]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef]
- Springer, S.A.; Gagneux, P. Glycan evolution in response to collaboration, conflict, and constraint. J. Biol. Chem. 2013, 288, 6904–6911. [Google Scholar] [CrossRef]
- Hart, G.W.; Varki, A. Future directions in glycosciences. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestergard, J.H., et al., Eds.; Cold Spring Harbor: Cold Spring Harbor, NY, USA, 2015; pp. 761–768. [Google Scholar]
- Meezan, E.; Wu, H.C.; Black, P.H.; Robbins, P.W. Comparative studies on the carbohydrate-containing membrane components of normal and virus-transformed mouse fibroblasts. Ii. Separation of glycoproteins and glycopeptides by sephadex chromatography. Biochemistry 1969, 8, 2518–2524. [Google Scholar] [CrossRef]
- Feizi, T. Carbohydrate antigens in human cancer. Cancer Surv. 1985, 4, 245–269. [Google Scholar]
- Hauselmann, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef]
- Munkley, J.; Elliott, D.J. Hallmarks of glycosylation in cancer. Oncotarget 2016, 7, 35478–35489. [Google Scholar] [CrossRef]
- Varki, A.; Kannagi, R.; Toole, B.; Stanley, P. Glycosylation changes in cancer. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestergard, J.H., et al., Eds.; Cold Spring Harbor: Cold Spring Harbor, NY, USA, 2015; pp. 597–609. [Google Scholar]
- Hammarstrom, S. The carcinoembryonic antigen (cea) family: Structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999, 9, 67–81. [Google Scholar] [CrossRef]
- Moss, E.L.; Hollingworth, J.; Reynolds, T.M. The role of ca125 in clinical practice. J. Clin. Pathol. 2005, 58, 308–312. [Google Scholar] [CrossRef]
- Gilgunn, S.; Conroy, P.J.; Saldova, R.; Rudd, P.M.; O’Kennedy, R.J. Aberrant psa glycosylation—A sweet predictor of prostate cancer. Nat. Rev. Urol. 2013, 10, 99–107. [Google Scholar] [CrossRef]
- Munkley, J.; Mills, I.G.; Elliott, D.J. The role of glycans in the development and progression of prostate cancer. Nat. Rev. Urol. 2016, 13, 324–333. [Google Scholar] [CrossRef]
- Tkac, J.; Bertok, T.; Hires, M.; Jane, E.; Lorencova, L.; Kasak, P. Glycomics of prostate cancer: Updates. Expert Rev. Proteom. 2018, 16, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Drake, R.R.; Jones, E.E.; Powers, T.W.; Nyalwidhe, J.O. Chapter ten—Altered glycosylation in prostate cancer. Adv. Cancer Res. 2015, 126, 345–382. [Google Scholar]
- Barry, M.J. Screening for prostate cancer—The controversy that refuses to die. N. Engl. J. Med. 2009, 360, 1351–1354. [Google Scholar] [CrossRef] [PubMed]
- Etzioni, R.; Penson, D.F.; Legler, J.M.; di Tommaso, D.; Boer, R.; Gann, P.H.; Feuer, E.J. Overdiagnosis due to prostate-specific antigen screening: Lessons from U.S. Prostate cancer incidence trends. J. Natl. Cancer Inst. 2002, 94, 981–990. [Google Scholar] [CrossRef]
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef]
- Nordstrom, T.; Vickers, A.; Assel, M.; Lilja, H.; Gronberg, H.; Eklund, M. Comparison between the four-kallikrein panel and prostate health index for predicting prostate cancer. Eur. Urol. 2015, 68, 139–146. [Google Scholar] [CrossRef]
- Prensner, J.R.; Rubin, M.A.; Wei, J.T.; Chinnaiyan, A.M. Beyond psa: The next generation of prostate cancer biomarkers. Sci. Transl. Med. 2012, 4, 127rv3. [Google Scholar] [CrossRef] [PubMed]
- Shariat, S.F.; Semjonow, A.; Lilja, H.; Savage, C.; Vickers, A.J.; Bjartell, A. Tumor markers in prostate cancer i: Blood-based markers. Acta Oncol. 2011, 50 (Suppl. 1), 61–75. [Google Scholar] [CrossRef]
- Sountoulides, P.; Moutzouris, G. Prostate-specific antigen screening, why have the guidelines changed? Expert Rev. Anticancer Ther. 2014, 14, 1277–1281. [Google Scholar] [CrossRef]
- Srinivasan, S.; Stephens, C.; Wilson, E.; Panchadsaram, J.; DeVoss, K.; Koistinen, H.; Stenman, U.H.; Brooks, M.N.; Practical, C.; Buckle, A.M.; et al. Prostate cancer risk-associated single-nucleotide polymorphism affects prostate-specific antigen glycosylation and its function. Clin. Chem. 2018, 65, e1–e9. [Google Scholar] [CrossRef]
- Llop, E.; Ferrer-Batalle, M.; Barrabes, S.; Guerrero, P.E.; Ramirez, M.; Saldova, R.; Rudd, P.M.; Aleixandre, R.N.; Comet, J.; de Llorens, R.; et al. Improvement of prostate cancer diagnosis by detecting psa glycosylation-specific changes. Theranostics 2016, 6, 1190–1204. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Batalle, M.; Llop, E.; Ramirez, M.; Aleixandre, R.N.; Saez, M.; Comet, J.; de Llorens, R.; Peracaula, R. Comparative study of blood-based biomarkers, alpha2,3-sialic acid psa and phi, for high-risk prostate cancer detection. Int. J. Mol. Sci. 2017, 18, 845. [Google Scholar] [CrossRef]
- Ishikawa, T.; Yoneyama, T.; Tobisawa, Y.; Hatakeyama, S.; Kurosawa, T.; Nakamura, K.; Narita, S.; Mitsuzuka, K.; Duivenvoorden, W.; Pinthus, J.H.; et al. An automated micro-total immunoassay system for measuring cancer-associated alpha2,3-linked sialyl n-glycan-carrying prostate-specific antigen may improve the accuracy of prostate cancer diagnosis. Int. J. Mol. Sci. 2017, 18, 470. [Google Scholar] [CrossRef]
- Vajaria, B.N.; Patel, K.R.; Begum, R.; Patel, P.S. Sialylation: An avenue to target cancer cells. Pathol. Oncol. Res. 2016, 22, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J. The role of sialyl-tn in cancer. Int. J. Mol. Sci. 2016, 17, 275. [Google Scholar] [CrossRef]
- Baldus, S.E.; Zirbes, T.K.; Monig, S.P.; Engel, S.; Monaca, E.; Rafiqpoor, K.; Hanisch, F.G.; Hanski, C.; Thiele, J.; Pichlmaier, H.; et al. Histopathological subtypes and prognosis of gastric cancer are correlated with the expression of mucin-associated sialylated antigens: Sialosyl-lewis(a), sialosyl-lewis(x) and sialosyl-tn. Tumour Biol. 1998, 19, 445–453. [Google Scholar] [CrossRef]
- Zhang, Z.; Wuhrer, M.; Holst, S. Serum sialylation changes in cancer. Glycoconj. J. 2018, 35, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J. The glycosylation landscape of pancreatic cancer (review). Oncol. Lett. 2019, 17, 2569–2575. [Google Scholar] [CrossRef]
- Michalakis, K.; Ilias, I.; Triantafyllou, A.; Polymeris, A.; Kastriotis, I.; Chairakaki, A.D.; Savopoulos, C. Detection of prostate cancer by sialic acid level in patients with non-diagnostic levels of prostate-specific antigen. Maturitas 2012, 73, 325–330. [Google Scholar] [CrossRef]
- El Melegy, N.T.; Aboulella, H.A.; Abul-Fadl, A.M.; Mohamed, N.A. Potential biomarkers for differentiation of benign prostatic hyperplasia and prostate cancer. Br. J. Biomed. Sci. 2010, 67, 109–112. [Google Scholar] [CrossRef]
- Saldova, R.; Fan, Y.; Fitzpatrick, J.M.; Watson, R.W.; Rudd, P.M. Core fucosylation and alpha2-3 sialylation in serum n-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology 2011, 21, 195–205. [Google Scholar] [CrossRef]
- Ohyama, C.; Hosono, M.; Nitta, K.; Oh-eda, M.; Yoshikawa, K.; Habuchi, T.; Arai, Y.; Fukuda, M. Carbohydrate structure and differential binding of prostate specific antigen to maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology 2004, 14, 671–679. [Google Scholar] [CrossRef]
- Zhang, C.; Yan, L.; Song, H.; Ma, Z.; Chen, D.; Yang, F.; Fang, L.; Li, Z.; Li, K.; Li, D.; et al. Elevated serum sialic acid levels predict prostate cancer as well as bone metastases. J. Cancer 2019, 10, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, S.T.; Bertozzi, C.R. Imaging the glycome. Proc. Natl. Acad. Sci. USA 2009, 106, 12–17. [Google Scholar] [CrossRef]
- Yang, L.; Nyalwidhe, J.O.; Guo, S.; Drake, R.R.; Semmes, O.J. Targeted identification of metastasis-associated cell-surface sialoglycoproteins in prostate cancer. Mol. Cell. Proteom. 2011, 10, M110-007294. [Google Scholar] [CrossRef] [PubMed]
- Spiciarich, D.R.; Nolley, R.; Maund, S.L.; Purcell, S.C.; Herschel, J.; Iavarone, A.T.; Peehl, D.M.; Bertozzi, C.R. Bioorthogonal labeling of human prostate cancer tissue slice cultures for glycoproteomics. Angew. Chem. Int. Ed. Engl. 2017, 56, 8992–8997. [Google Scholar] [CrossRef]
- Culig, Z.; Hittmair, A.; Hobisch, A.; Bartsch, G.; Klocker, H.; Pai, L.H.; Pastan, I. Expression of lewis carbohydrate antigens in metastatic lesions from human prostatic carcinoma. Prostate 1998, 36, 162–167. [Google Scholar] [CrossRef]
- Jorgensen, T.; Berner, A.; Kaalhus, O.; Tveter, K.J.; Danielsen, H.E.; Bryne, M. Up-regulation of the oligosaccharide sialyl lewisx: A new prognostic parameter in metastatic prostate cancer. Cancer Res. 1995, 55, 1817–1819. [Google Scholar] [PubMed]
- Khabaz, M.N.; McClure, J.; McClure, S.; Stoddart, R.W. Glycophenotype of prostatic carcinomas. Folia Histochem. Cytobiol. 2010, 48, 637–645. [Google Scholar]
- Okamoto, T.; Yoneyama, M.S.; Hatakeyama, S.; Mori, K.; Yamamoto, H.; Koie, T.; Saitoh, H.; Yamaya, K.; Funyu, T.; Fukuda, M.; et al. Core2 o-glycan-expressing prostate cancer cells are resistant to nk cell immunity. Mol. Med. Rep. 2013, 7, 359–364. [Google Scholar] [CrossRef]
- Natoni, A.; Macauley, M.S.; O’Dwyer, M.E. Targeting selectins and their ligands in cancer. Front. Oncol. 2016, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- Dimitroff, C.J.; Lechpammer, M.; Long-Woodward, D.; Kutok, J.L. Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by e-selectin. Cancer Res. 2004, 64, 5261–5269. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.; Ivetic, A.; Grigoriadis, A.; QiZe, D.; Burford, B.; Sproviero, D.; Picco, G.; Gillett, C.; Papp, S.L.; Schaffer, L.; et al. Selectin ligand sialyl-lewis x antigen drives metastasis of hormone-dependent breast cancers. Cancer Res. 2011, 71, 7683–7693. [Google Scholar] [CrossRef]
- Barthel, S.R.; Wiese, G.K.; Cho, J.; Opperman, M.J.; Hays, D.L.; Siddiqui, J.; Pienta, K.J.; Furie, B.; Dimitroff, C.J. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc. Natl. Acad. Sci. USA 2009, 106, 19491–19496. [Google Scholar] [CrossRef]
- Chen, Z.; Gulzar, Z.G.; St Hill, C.A.; Walcheck, B.; Brooks, J.D. Increased expression of gcnt1 is associated with altered o-glycosylation of psa, pap, and muc1 in human prostate cancers. Prostate 2014, 74, 1059–1067. [Google Scholar] [CrossRef]
- Myers, R.B.; Meredith, R.F.; Schlom, J.; LoBuglio, A.F.; Bueschen, A.J.; Wheeler, R.H.; Stockard, C.R.; Grizzle, W.E. Tumor associated glycoprotein-72 is highly expressed in prostatic adenocarcinomas. J. Urol. 1994, 152, 243–246. [Google Scholar] [CrossRef]
- Genega, E.M.; Hutchinson, B.; Reuter, V.E.; Gaudin, P.B. Immunophenotype of high-grade prostatic adenocarcinoma and urothelial carcinoma. Mod. Pathol. 2000, 13, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J.; Oltean, S.; Vodak, D.; Wilson, B.T.; Livermore, K.E.; Zhou, Y.; Star, E.; Floros, V.I.; Johannessen, B.; Knight, B.; et al. The androgen receptor controls expression of the cancer-associated stn antigen and cell adhesion through induction of st6galnac1 in prostate cancer. Oncotarget 2015, 6, 34358–34374. [Google Scholar] [CrossRef]
- Munkley, J.; Elliott, D.J. Sugars and cell adhesion: The role of st6galnac1 in prostate cancer progression. Cancer Cell Microenviron. 2016, 3, e1174. [Google Scholar]
- Lima, L.; Neves, M.; Oliveira, M.I.; Dieguez, L.; Freitas, R.; Azevedo, R.; Gaiteiro, C.; Soares, J.; Ferreira, D.; Peixoto, A.; et al. Sialyl-tn identifies muscle-invasive bladder cancer basal and luminal subtypes facing decreased survival, being expressed by circulating tumor cells and metastases. Urol. Oncol. 2017, 35, 675 e671–675 e678. [Google Scholar] [CrossRef]
- Neves, M.; Azevedo, R.; Lima, L.; Oliveira, M.I.; Peixoto, A.; Ferreira, D.; Soares, J.; Fernandes, E.; Gaiteiro, C.; Palmeira, C.; et al. Exploring sialyl-tn expression in microfluidic-isolated circulating tumour cells: A novel biomarker and an analytical tool for precision oncology applications. New Biotechnol. 2019, 49, 77–87. [Google Scholar] [CrossRef]
- Eavarone, D.A.; Al-Alem, L.; Lugovskoy, A.; Prendergast, J.M.; Nazer, R.I.; Stein, J.N.; Dransfield, D.T.; Behrens, J.; Rueda, B.R. Humanized anti-sialyl-tn antibodies for the treatment of ovarian carcinoma. PLoS ONE 2018, 13, e0201314. [Google Scholar] [CrossRef]
- Prendergast, J.M.; Galvao da Silva, A.P.; Eavarone, D.A.; Ghaderi, D.; Zhang, M.; Brady, D.; Wicks, J.; DeSander, J.; Behrens, J.; Rueda, B.R. Novel anti-sialyl-tn monoclonal antibodies and antibody-drug conjugates demonstrate tumor specificity and anti-tumor activity. mAbs 2017, 9, 615–627. [Google Scholar] [CrossRef]
- Totten, S.M.; Adusumilli, R.; Kullolli, M.; Tanimoto, C.; Brooks, J.D.; Mallick, P.; Pitteri, S.J. Multi-lectin affinity chromatography and quantitative proteomic analysis reveal differential glycoform levels between prostate cancer and benign prostatic hyperplasia sera. Sci. Rep. 2018, 8, 6509. [Google Scholar] [CrossRef]
- Kyselova, Z.; Mechref, Y.; Al Bataineh, M.M.; Dobrolecki, L.E.; Hickey, R.J.; Vinson, J.; Sweeney, C.J.; Novotny, M.V. Alterations in the serum glycome due to metastatic prostate cancer. J. Proteome Res. 2007, 6, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Shimomura, M.; Uemura, M.; Nakata, W.; Sato, M.; Nagahara, A.; Nakai, Y.; Takamatsu, S.; Miyoshi, E.; Nonomura, N. Serum fucosylated haptoglobin as a novel prognostic biomarker predicting high-gleason prostate cancer. Prostate 2014, 74, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Wang, X.; Yang, W.; Toghi Eshghi, S.; Sun, S.; Hoti, N.; Chen, L.; Yang, S.; Pasay, J.; Rubin, A.; et al. Integrated proteomic and glycoproteomic analyses of prostate cancer cells reveal glycoprotein alteration in protein abundance and glycosylation. Mol. Cell. Proteom. 2015, 14, 2753–2763. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Gao, J.; Han, C.; Zhang, X.; Liu, H.; Ma, L.; Sun, X.; Yu, W. O-glcnacylation is increased in prostate cancer tissues and enhances malignancy of prostate cancer cells. Mol. Med. Rep. 2014, 10, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Kamigaito, T.; Okaneya, T.; Kawakubo, M.; Shimojo, H.; Nishizawa, O.; Nakayama, J. Overexpression of o-glcnac by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis. 2014, 17, 18–22. [Google Scholar] [CrossRef]
- Kaushik, A.K.; Shojaie, A.; Panzitt, K.; Sonavane, R.; Venghatakrishnan, H.; Manikkam, M.; Zaslavsky, A.; Putluri, V.; Vasu, V.T.; Zhang, Y.; et al. Inhibition of the hexosamine biosynthetic pathway promotes castration-resistant prostate cancer. Nat. Commun. 2016, 7, 11612. [Google Scholar] [CrossRef]
- Lange, T.; Ullrich, S.; Muller, I.; Nentwich, M.F.; Stubke, K.; Feldhaus, S.; Knies, C.; Hellwinkel, O.J.; Vessella, R.L.; Abramjuk, C.; et al. Human prostate cancer in a clinically relevant xenograft mouse model: Identification of beta(1,6)-branched oligosaccharides as a marker of tumor progression. Clin. Cancer Res. 2012, 18, 1364–1373. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Tobisawa, Y.; Hatakeyama, S.; Ohashi, T.; Tanaka, M.; Narita, S.; Koie, T.; Habuchi, T.; Nishimura, S.; Ohyama, C.; et al. Serum tri- and tetra-antennary n-glycan is a potential predictive biomarker for castration-resistant prostate cancer. Prostate 2014, 74, 1521–1529. [Google Scholar] [CrossRef]
- Wang, D.; Dafik, L.; Nolley, R.; Huang, W.; Wolfinger, R.D.; Wang, L.X.; Peehl, D.M. Anti-oligomannose antibodies as potential serum biomarkers of aggressive prostate cancer. Drug Dev. Res. 2013, 74, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, A.; Kitazume, S.; Kizuka, Y.; Nakajima, K.; Oka, R.; Fujinawa, R.; Korekane, H.; Yamaguchi, Y.; Wada, Y.; Taniguchi, N. The absence of core fucose up-regulates gnt-iii and wnt target genes: A possible mechanism for an adaptive response in terms of glycan function. J. Biol. Chem. 2014, 289, 11704–11714. [Google Scholar] [CrossRef]
- Wang, X.; Chen, J.; Li, Q.K.; Peskoe, S.B.; Zhang, B.; Choi, C.; Platz, E.A.; Zhang, H. Overexpression of alpha (1,6) fucosyltransferase associated with aggressive prostate cancer. Glycobiology 2014, 24, 935–944. [Google Scholar] [CrossRef] [PubMed]
- Hoti, N.; Yang, S.; Hu, Y.; Shah, P.; Haffner, M.C.; Zhang, H. Overexpression of alpha (1,6) fucosyltransferase in the development of castration-resistant prostate cancer cells. Prostate Cancer Prostatic Dis. 2018, 21, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Hayashi, T.; Matsuzaki, K.; Nakata, W.; Masuda, M.; Kawashima, A.; Ujike, T.; Nagahara, A.; Tsuchiya, M.; Kobayashi, Y.; et al. Decreased fucosylated psa as a urinary marker for high gleason score prostate cancer. Oncotarget 2016, 7, 56643–56649. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, W.; Hu, Y.; Hoti, N.; Liu, Y.; Shah, P.; Sun, S.; Clark, D.; Thomas, S.; Zhang, H. Site-specific fucosylation analysis identifying glycoproteins associated with aggressive prostate cancer cell lines using tandem affinity enrichments of intact glycopeptides followed by mass spectrometry. Anal. Chem. 2017, 89, 7623–7630. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of o-glcnac. J. Cell Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef]
- Hanover, J.A.; Krause, M.W.; Love, D.C. Bittersweet memories: Linking metabolism to epigenetics through o-glcnacylation. Nat. Rev. Mol. Cell Biol. 2012, 13, 312–321. [Google Scholar] [CrossRef]
- Fardini, Y.; Dehennaut, V.; Lefebvre, T.; Issad, T. O-glcnacylation: A new cancer hallmark? Front. Endocrinol. 2013, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Hanover, J.A.; Chen, W.; Bond, M.R. O-glcnac in cancer: An oncometabolism-fueled vicious cycle. J. Bioenergy Biomembr. 2018, 50, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Gil, M.; Pierce, A.; Perez-Cervera, Y.; Zenteno, E.; Lefebvre, T. Ogt: A short overview of an enzyme standing out from usual glycosyltransferases. Biochem. Soc. Trans. 2017, 45, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Chiaradonna, F.; Ricciardiello, F.; Palorini, R. The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic rewiring. Cells 2018, 7, 53. [Google Scholar] [CrossRef]
- Itkonen, H.M.; Minner, S.; Guldvik, I.J.; Sandmann, M.J.; Tsourlakis, M.C.; Berge, V.; Svindland, A.; Schlomm, T.; Mills, I.G. O-glcnac transferase integrates metabolic pathways to regulate the stability of c-myc in human prostate cancer cells. Cancer Res. 2013, 73, 5277–5287. [Google Scholar] [CrossRef]
- Itkonen, H.M.; Engedal, N.; Babaie, E.; Luhr, M.; Guldvik, I.J.; Minner, S.; Hohloch, J.; Tsourlakis, M.C.; Schlomm, T.; Mills, I.G. Uap1 is overexpressed in prostate cancer and is protective against inhibitors of n-linked glycosylation. Oncogene 2014, 34, 3744. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.W.; Granovsky, M.; Warren, C.E. Glycoprotein glycosylation and cancer progression. Biochim. Biophys. Acta 1999, 1473, 21–34. [Google Scholar] [CrossRef]
- Taniguchi, N.; Kizuka, Y. Glycans and cancer: Role of n-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015, 126, 11–51. [Google Scholar]
- Kawahara, R.; Ortega, F.; Rosa-Fernandes, L.; Guimaraes, V.; Quina, D.; Nahas, W.; Schwammle, V.; Srougi, M.; Leite, K.R.M.; Thaysen-Andersen, M.; et al. Distinct urinary glycoprotein signatures in prostate cancer patients. Oncotarget 2018, 9, 33077–33097. [Google Scholar] [CrossRef]
- Bhat, G.; Hothpet, V.R.; Lin, M.F.; Cheng, P.W. Shifted golgi targeting of glycosyltransferases and alpha-mannosidase ia from giantin to gm130-grasp65 results in formation of high mannose n-glycans in aggressive prostate cancer cells. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 2891–2901. [Google Scholar] [CrossRef]
- Newsom-Davis, T.E.; Wang, D.; Steinman, L.; Chen, P.F.; Wang, L.X.; Simon, A.K.; Screaton, G.R. Enhanced immune recognition of cryptic glycan markers in human tumors. Cancer Res. 2009, 69, 2018–2025. [Google Scholar] [CrossRef]
- Wang, D. N-glycan cryptic antigens as active immunological targets in prostate cancer patients. J. Proteom. Bioinform. 2012, 5, 090–095. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Herzenberg, L.; Peehl, D.; Herzenberg, L. Prostate Cancer Glycan Markers and Auto-Antibody Signatures. U.S. Patent No. 7,981,625, 19 July 2011. [Google Scholar]
- Handerson, T.; Pawelek, J.M. Beta1,6-branched oligosaccharides and coarse vesicles: A common, pervasive phenotype in melanoma and other human cancers. Cancer Res. 2003, 63, 5363–5369. [Google Scholar]
- Nonaka, M.; Fukuda, M.N.; Gao, C.; Li, Z.; Zhang, H.; Greene, M.I.; Peehl, D.M.; Feizi, T.; Fukuda, M. Determination of carbohydrate structure recognized by prostate-specific f77 monoclonal antibody through expression analysis of glycosyltransferase genes. J. Biol. Chem. 2014, 289, 16478–16486. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, H.; Wang, Q.; Lal, P.; Carroll, A.M.; de la Llera-Moya, M.; Xu, X.; Greene, M.I. Suppression of human prostate tumor growth by a unique prostate-specific monoclonal antibody f77 targeting a glycolipid marker. Proc. Natl. Acad. Sci. USA 2010, 107, 732–737. [Google Scholar] [CrossRef]
- Chen, X.; Nagai, Y.; Zhu, Z.; Ruan, H.; Peehl, D.M.; Greene, M.I.; Zhang, H. A spliced form of cd44 expresses the unique glycan that is recognized by the prostate cancer specific antibody f77. Oncotarget 2018, 9, 3631–3640. [Google Scholar] [CrossRef] [PubMed]
- Daniotti, J.L.; Lardone, R.D.; Vilcaes, A.A. Dysregulated expression of glycolipids in tumor cells: From negative modulator of anti-tumor immunity to promising targets for developing therapeutic agents. Front. Oncol. 2015, 5, 300. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Muthugounder, S.; Presser, N.; Selvan, S.R.; Portoukalian, J.; Brosman, S.; Morton, D.L. Gangliosides of organ-confined versus metastatic androgen-receptor-negative prostate cancer. Biochem. Biophys. Res. Commun. 2004, 324, 154–165. [Google Scholar] [CrossRef]
- Hatano, K.; Miyamoto, Y.; Mori, M.; Nimura, K.; Nakai, Y.; Nonomura, N.; Kaneda, Y. Androgen-regulated transcriptional control of sialyltransferases in prostate cancer cells. PLoS ONE 2012, 7, e31234. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V.; Sanderson, R.D. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 2011, 15, 1013–1031. [Google Scholar] [CrossRef]
- Edwards, I.J. Proteoglycans in prostate cancer. Nat. Rev. Urol. 2012, 9, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Ricciardelli, C.; Mayne, K.; Sykes, P.J.; Raymond, W.A.; McCaul, K.; Marshall, V.R.; Horsfall, D.J. Elevated levels of versican but not decorin predict disease progression in early-stage prostate cancer. Clin. Cancer Res. 1998, 4, 963–971. [Google Scholar]
- Jacobsen, F.; Kraft, J.; Schroeder, C.; Hube-Magg, C.; Kluth, M.; Lang, D.S.; Simon, R.; Sauter, G.; Izbicki, J.R.; Clauditz, T.S.; et al. Up-regulation of biglycan is associated with poor prognosis and pten deletion in patients with prostate cancer. Neoplasia 2017, 19, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, B.; Alghezi, D.A.; Cattermole, C.; Beresford, M.; Bowen, R.; Mitchard, J.; Chalmers, A.D. A subset of high gleason grade prostate carcinomas contain a large burden of prostate cancer syndecan-1 positive stromal cells. Prostate 2017, 77, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Szarvas, T.; Reis, H.; Vom Dorp, F.; Tschirdewahn, S.; Niedworok, C.; Nyirady, P.; Schmid, K.W.; Rubben, H.; Kovalszky, I. Soluble syndecan-1 (sdc1) serum level as an independent pre-operative predictor of cancer-specific survival in prostate cancer. Prostate 2016, 76, 977–985. [Google Scholar] [CrossRef]
- Sakko, A.J.; Butler, M.S.; Byers, S.; Reinboth, B.J.; Stahl, J.; Kench, J.G.; Horvath, L.G.; Sutherland, R.L.; Stricker, P.D.; Henshall, S.M.; et al. Immunohistochemical level of unsulfated chondroitin disaccharides in the cancer stroma is an independent predictor of prostate cancer relapse. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2488–2497. [Google Scholar] [CrossRef]
- Sakko, A.J.; Ricciardelli, C.; Mayne, K.; Suwiwat, S.; LeBaron, R.G.; Marshall, V.R.; Tilley, W.D.; Horsfall, D.J. Modulation of prostate cancer cell attachment to matrix by versican. Cancer Res. 2003, 63, 4786–4791. [Google Scholar] [PubMed]
- Ricciardelli, C.; Russell, D.L.; Ween, M.P.; Mayne, K.; Suwiwat, S.; Byers, S.; Marshall, V.R.; Tilley, W.D.; Horsfall, D.J. Formation of hyaluronan- and versican-rich pericellular matrix by prostate cancer cells promotes cell motility. J. Biol. Chem. 2007, 282, 10814–10825. [Google Scholar] [CrossRef]
- Hu, Y.; Sun, H.; Owens, R.T.; Wu, J.; Chen, Y.Q.; Berquin, I.M.; Perry, D.; O’Flaherty, J.T.; Edwards, I.J. Decorin suppresses prostate tumor growth through inhibition of epidermal growth factor and androgen receptor pathways. Neoplasia 2009, 11, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Henke, A.; Grace, O.C.; Ashley, G.R.; Stewart, G.D.; Riddick, A.C.; Yeun, H.; O’Donnell, M.; Anderson, R.A.; Thomson, A.A. Stromal expression of decorin, semaphorin6d, sparc, sprouty1 and tsukushi in developing prostate and decreased levels of decorin in prostate cancer. PLoS ONE 2012, 7, e42516. [Google Scholar] [CrossRef]
- Coulson-Thomas, V.J.; Coulson-Thomas, Y.M.; Gesteira, T.F.; Andrade de Paula, C.A.; Carneiro, C.R.; Ortiz, V.; Toma, L.; Kao, W.W.; Nader, H.B. Lumican expression, localization and antitumor activity in prostate cancer. Exp. Cell Res. 2013, 319, 967–981. [Google Scholar] [CrossRef]
- Datta, M.W.; Hernandez, A.M.; Schlicht, M.J.; Kahler, A.J.; DeGueme, A.M.; Dhir, R.; Shah, R.B.; Farach-Carson, C.; Barrett, A.; Datta, S. Perlecan, a candidate gene for the capb locus, regulates prostate cancer cell growth via the sonic hedgehog pathway. Mol. Cancer 2006, 5, 9. [Google Scholar] [CrossRef]
- Grindel, B.; Li, Q.; Arnold, R.; Petros, J.; Zayzafoon, M.; Muldoon, M.; Stave, J.; Chung, L.W.; Farach-Carson, M.C. Perlecan/hspg2 and matrilysin/mmp-7 as indices of tissue invasion: Tissue localization and circulating perlecan fragments in a cohort of 288 radical prostatectomy patients. Oncotarget 2016, 7, 10433–10447. [Google Scholar] [PubMed]
- Fujii, T.; Shimada, K.; Tatsumi, Y.; Tanaka, N.; Fujimoto, K.; Konishi, N. Syndecan-1 up-regulates microrna-331-3p and mediates epithelial-to-mesenchymal transition in prostate cancer. Mol. Carcinog. 2015. [Google Scholar]
- Shimada, K.; Anai, S.; Fujii, T.; Tanaka, N.; Fujimoto, K.; Konishi, N. Syndecan-1 (cd138) contributes to prostate cancer progression by stabilizing tumour-initiating cells. J. Pathol. 2013, 231, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, A.H.; Alalawi, Z.; Mirandola, L.; Rakhshanda, R.; Dahlbeck, S.; Nguyen, D.; Jenkins, M.; Grizzi, F.; Cobos, E.; Figueroa, J.A.; et al. Galectins in cancer: Carcinogenesis, diagnosis and therapy. Ann. Transl. Med. 2014, 2, 88. [Google Scholar] [PubMed]
- Laderach, D.J.; Gentilini, L.D.; Giribaldi, L.; Delgado, V.C.; Nugnes, L.; Croci, D.O.; Al Nakouzi, N.; Sacca, P.; Casas, G.; Mazza, O.; et al. A unique galectin signature in human prostate cancer progression suggests galectin-1 as a key target for treatment of advanced disease. Cancer Res. 2013, 73, 86–96. [Google Scholar] [CrossRef]
- Nakajima, K.; Kho, D.H.; Yanagawa, T.; Harazono, Y.; Hogan, V.; Chen, W.; Ali-Fehmi, R.; Mehra, R.; Raz, A. Galectin-3 cleavage alters bone remodeling: Different outcomes in breast and prostate cancer skeletal metastasis. Cancer Res. 2016, 76, 1391–1402. [Google Scholar] [CrossRef]
- Dondoo, T.O.; Fukumori, T.; Daizumoto, K.; Fukawa, T.; Kohzuki, M.; Kowada, M.; Kusuhara, Y.; Mori, H.; Nakatsuji, H.; Takahashi, M.; et al. Galectin-3 is implicated in tumor progression and resistance to anti-androgen drug through regulation of androgen receptor signaling in prostate cancer. Anticancer Res. 2017, 37, 125–134. [Google Scholar] [CrossRef]
- Knapp, J.S.; Lokeshwar, S.D.; Vogel, U.; Hennenlotter, J.; Schwentner, C.; Kramer, M.W.; Stenzl, A.; Merseburger, A.S. Galectin-3 expression in prostate cancer and benign prostate tissues: Correlation with biochemical recurrence. World J. Urol. 2013, 31, 351–358. [Google Scholar] [CrossRef]
- Payton, S. Prostate cancer: ‘Galectin signature’ reveals gal-1 as key player in angiogenesis. Nat. Rev. Urol. 2012, 9, 667. [Google Scholar] [CrossRef]
- Shih, T.C.; Liu, R.; Wu, C.T.; Li, X.; Xiao, W.; Deng, X.; Kiss, S.; Wang, T.; Chen, X.J.; Carney, R.; et al. Targeting galectin-1 impairs castration-resistant prostate cancer progression and invasion. Clin. Cancer Res. 2018, 24, 4319–4331. [Google Scholar] [CrossRef]
- Wang, Y.; Nangia-Makker, P.; Tait, L.; Balan, V.; Hogan, V.; Pienta, K.J.; Raz, A. Regulation of prostate cancer progression by galectin-3. Am. J. Pathol. 2009, 174, 1515–1523. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Tzeng, S.F.; Chao, T.K.; Tsai, C.Y.; Yang, Y.C.; Lee, M.T.; Hwang, J.J.; Chou, Y.C.; Tsai, M.H.; Cha, T.L.; et al. Metastatic progression of prostate cancer is mediated by autonomous binding of galectin-4-o-glycan to cancer cells. Cancer Res. 2016, 76, 5756–5767. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, S.F.; Tsai, C.H.; Chao, T.K.; Chou, Y.C.; Yang, Y.C.; Tsai, M.H.; Cha, T.L.; Hsiao, P.W. O-glycosylation-mediated signaling circuit drives metastatic castration-resistant prostate cancer. FASEB J. 2018, 32, 6869–6882. [Google Scholar] [CrossRef]
- Gentilini, L.D.; Jaworski, F.M.; Tiraboschi, C.; Perez, I.G.; Kotler, M.L.; Chauchereau, A.; Laderach, D.J.; Compagno, D. Stable and high expression of galectin-8 tightly controls metastatic progression of prostate cancer. Oncotarget 2017, 8, 44654–44668. [Google Scholar] [CrossRef] [PubMed]
- Munkley, J. Glycosylation is a global target for androgen control in prostate cancer cells. Endocr. Relat. Cancer 2017, 24, R49–R64. [Google Scholar] [CrossRef]
- Munkley, J.; Vodak, D.; Livermore, K.E.; James, K.; Wilson, B.T.; Knight, B.; McCullagh, P.; McGrath, J.; Crundwell, M.; Harries, L.W.; et al. Glycosylation is an androgen-regulated process essential for prostate cancer cell viability. EBioMedicine 2016, 8, 103–116. [Google Scholar] [CrossRef]
- Sato, T.; Yoneyama, T.; Tobisawa, Y.; Hatakeyama, S.; Yamamoto, H.; Kojima, Y.; Mikami, J.; Mori, K.; Hashimoto, Y.; Koie, T.; et al. Core 2 beta-1, 6-n-acetylglucosaminyltransferase-1 expression in prostate biopsy specimen is an indicator of prostate cancer aggressiveness. Biochem. Biophys. Res. Commun. 2016, 470, 150–156. [Google Scholar] [CrossRef]
- Hagisawa, S.; Ohyama, C.; Takahashi, T.; Endoh, M.; Moriya, T.; Nakayama, J.; Arai, Y.; Fukuda, M. Expression of core 2 beta1,6-n-acetylglucosaminyltransferase facilitates prostate cancer progression. Glycobiology 2005, 15, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.; Fan, B.; Zhao, Y.; Zhang, H.; Wang, L.; Yu, X.; Yuan, Q.; Yang, D.; Wang, S. St6gal-i overexpression facilitates prostate cancer progression via the pi3k/akt/gsk-3beta/beta-catenin signaling pathway. Oncotarget 2016, 7, 65374–65388. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guillebon, A.D.; Hsu, J.W.; Barthel, S.R.; Dimitroff, C.J.; Lee, Y.F.; King, M.R. Human fucosyltransferase 6 enables prostate cancer metastasis to bone. Br. J. Cancer 2013, 109, 3014–3022. [Google Scholar] [CrossRef]
- Livermore, K.E.; Munkley, J.; Elliott, D.J. Androgen receptor and prostate cancer. AIMS Mol. Sci. 2016, 3, 280–299. [Google Scholar] [CrossRef]
- Sharp, A.; Coleman, I.; Yuan, W.; Sprenger, C.; Dolling, D.; Rodrigues, D.N.; Russo, J.W.; Figueiredo, I.; Bertan, C.; Seed, G.; et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Investig. 2019, 129, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Yoneyama, T.; Hatakeyama, S.; Mikami, J.; Sato, T.; Mori, K.; Hashimoto, Y.; Koie, T.; Ohyama, C.; Fukuda, M.; et al. Detection of core2 beta-1,6-n-acetylglucosaminyltransferase in post-digital rectal examination urine is a reliable indicator for extracapsular extension of prostate cancer. PLoS ONE 2015, 10, e0138520. [Google Scholar] [CrossRef]
- Albitar, M.; Ma, W.; Lund, L.; Albitar, F.; Diep, K.; Fritsche, H.A.; Shore, N. Predicting prostate biopsy results using a panel of plasma and urine biomarkers combined in a scoring system. J. Cancer 2016, 7, 297–303. [Google Scholar] [CrossRef]
- Ma, W.; Diep, K.; Fritsche, H.A.; Shore, N.; Albitar, M. Diagnostic and prognostic scoring system for prostate cancer using urine and plasma biomarkers. Genet. Test. Mol. Biomark. 2014, 18, 156–163. [Google Scholar] [CrossRef]
- Knuuttila, M.; Mehmood, A.; Huhtaniemi, R.; Yatkin, E.; Hakkinen, M.R.; Oksala, R.; Laajala, T.D.; Ryberg, H.; Handelsman, D.J.; Aittokallio, T.; et al. Antiandrogens reduce intratumoral androgen concentrations and induce androgen receptor expression in castration-resistant prostate cancer xenografts. Am. J. Pathol. 2018, 188, 216–228. [Google Scholar] [CrossRef]
- Mikami, J.; Tobisawa, Y.; Yoneyama, T.; Hatakeyama, S.; Mori, K.; Hashimoto, Y.; Koie, T.; Ohyama, C.; Fukuda, M. I-branching n-acetylglucosaminyltransferase regulates prostate cancer invasiveness by enhancing alpha5beta1 integrin signaling. Cancer Sci. 2016, 107, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Barfeld, S.J.; East, P.; Zuber, V.; Mills, I.G. Meta-analysis of prostate cancer gene expression data identifies a novel discriminatory signature enriched for glycosylating enzymes. BMC Med. Genom. 2014, 7, 513. [Google Scholar] [CrossRef]
- Tsui, K.H.; Chang, P.L.; Feng, T.H.; Chung, L.C.; Sung, H.C.; Juang, H.H. Evaluating the function of matriptase and n-acetylglucosaminyltransferase v in prostate cancer metastasis. Anticancer Res. 2008, 28, 1993–1999. [Google Scholar] [PubMed]
- Soung, Y.H.; Ford, S.; Zhang, V.; Chung, J. Exosomes in cancer diagnostics. Cancers 2017, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Drake, R.R.; Kislinger, T. The proteomics of prostate cancer exosomes. Expert Rev. Proteom. 2014, 11, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Royo, F.; Aizpurua-Olaizola, O.; Pazos, R.; Boons, G.J.; Reichardt, N.C.; Falcon-Perez, J.M. Glycosylation of extracellular vesicles: Current knowledge, tools and clinical perspectives. J. Extracell. Vesicles 2018, 7, 1442985. [Google Scholar] [CrossRef] [PubMed]
- Vlaeminck-Guillem, V. Extracellular vesicles in prostate cancer carcinogenesis, diagnosis, and management. Front. Oncol. 2018, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, J.Q.; Griffin, M.D. Getting to know the extracellular vesicle glycome. Mol. Biosyst. 2016, 12, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Nyalwidhe, J.O.; Betesh, L.R.; Powers, T.W.; Jones, E.E.; White, K.Y.; Burch, T.C.; Brooks, J.; Watson, M.T.; Lance, R.S.; Troyer, D.A.; et al. Increased bisecting n-acetylglucosamine and decreased branched chain glycans of n-linked glycoproteins in expressed prostatic secretions associated with prostate cancer progression. Proteom. Clin. Appl. 2013, 7, 677–689. [Google Scholar]
- Lopez Aguilar, A.; Meng, L.; Hou, X.; Li, W.; Moremen, K.W.; Wu, P. Sialyltransferase-based chemoenzymatic histology for the detection of n- and o-glycans. Bioconjugate Chem. 2018, 29, 1231–1239. [Google Scholar] [CrossRef]
- Powers, T.W.; Neely, B.A.; Shao, Y.; Tang, H.; Troyer, D.A.; Mehta, A.S.; Haab, B.B.; Drake, R.R. Maldi imaging mass spectrometry profiling of n-glycans in formalin-fixed paraffin embedded clinical tissue blocks and tissue microarrays. PLoS ONE 2014, 9, e106255. [Google Scholar] [CrossRef]
- Lemaire, R.; Desmons, A.; Tabet, J.C.; Day, R.; Salzet, M.; Fournier, I. Direct analysis and maldi imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res. 2007, 6, 1295–1305. [Google Scholar] [CrossRef]
- Drake, R.R.; West, C.A.; Mehta, A.S.; Angel, P.M. Maldi mass spectrometry imaging of n-linked glycans in tissues. Adv. Exp. Med. Biol. 2018, 1104, 59–76. [Google Scholar]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Vandekerkhove, G.; Chi, K.N.; Wyatt, A.W. Clinical utility of emerging liquid biomarkers in advanced prostate cancer. Cancer Genet. 2018, 228–229, 151–158. [Google Scholar] [CrossRef]
- Riaz, I.B.; Wang, L.; Kohli, M. Liquid biopsy approach in the management of prostate cancer. Transl. Res. 2018, 201, 60–70. [Google Scholar] [CrossRef]
- Gomella, L.G. The liquid biopsy for prostate cancer 25 years later. Can. J. Urol. 2017, 24, 8693–8694. [Google Scholar]
- Morrison, G.J.; Goldkorn, A. Development and application of liquid biopsies in metastatic prostate cancer. Curr. Oncol. Rep. 2018, 20, 35. [Google Scholar] [CrossRef]
- Fletcher, C. Ar-v7 liquid biopsy for treatment stratification in prostate cancer: How close are we? Curr. Opin. Urol. 2017, 27, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Valentino, A.; Reclusa, P.; Sirera, R.; Giallombardo, M.; Camps, C.; Pauwels, P.; Crispi, S.; Rolfo, C. Exosomal micrornas in liquid biopsies: Future biomarkers for prostate cancer. Clin. Transl. Oncol. 2017, 19, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Zhang, G.L.; Li, H.R.; Luo, J.D.; Li, Z.X.; Chen, G.M.; Yang, J. A panel of five circulating micrornas as potential biomarkers for prostate cancer. Prostate 2012, 72, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Haldrup, C.; Kosaka, N.; Ochiya, T.; Borre, M.; Hoyer, S.; Orntoft, T.F.; Sorensen, K.D. Profiling of circulating micrornas for prostate cancer biomarker discovery. Drug Deliv. Transl. Res. 2014, 4, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Groskopf, J.; Aubin, S.M.; Deras, I.L.; Blase, A.; Bodrug, S.; Clark, C.; Brentano, S.; Mathis, J.; Pham, J.; Meyer, T.; et al. Aptima pca3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 2006, 52, 1089–1095. [Google Scholar] [CrossRef]
- Salami, S.S.; Schmidt, F.; Laxman, B.; Regan, M.M.; Rickman, D.S.; Scherr, D.; Bueti, G.; Siddiqui, J.; Tomlins, S.A.; Wei, J.T.; et al. Combining urinary detection of tmprss2:Erg and pca3 with serum psa to predict diagnosis of prostate cancer. Urol. Oncol. 2013, 31, 566–571. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Aubin, S.M.; Siddiqui, J.; Lonigro, R.J.; Sefton-Miller, L.; Miick, S.; Williamsen, S.; Hodge, P.; Meinke, J.; Blase, A.; et al. Urine tmprss2:Erg fusion transcript stratifies prostate cancer risk in men with elevated serum psa. Sci. Transl. Med. 2011, 3, 94ra72. [Google Scholar] [CrossRef]
- Wu, T.; Giovannucci, E.; Welge, J.; Mallick, P.; Tang, W.Y.; Ho, S.M. Measurement of gstp1 promoter methylation in body fluids may complement psa screening: A meta-analysis. Br. J. Cancer 2011, 105, 65–73. [Google Scholar] [CrossRef]
- He, W.S.; Bishop, K.S. The potential use of cell-free-circulating-tumor DNA as a biomarker for prostate cancer. Expert Rev. Mol. Diagn. 2016, 16, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ding, M.; Xu, K.; Yang, C.; Mao, L.J. Exosomes in diagnosis and therapy of prostate cancer. Oncotarget 2017, 8, 97693–97700. [Google Scholar] [CrossRef]
- Sumanasuriya, S.; Omlin, A.G.; Armstrong, A.J.; Attard, G.; Chi, K.N.; Bevan, C.L.; Waugh, D.J.; Ijzerman, M.J.; De Laere, B.; Lolkema, M.P.; et al. Consensus statement on circulating biomarkers for advanced prostate cancer. Eur. Urol. Oncol. 2018, 1, 151–159. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer diagnosis using a liquid biopsy: Challenges and expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef]
- Ko, J.; Baldassano, S.N.; Loh, P.L.; Kording, K.; Litt, B.; Issadore, D. Machine learning to detect signatures of disease in liquid biopsies—A user’s guide. Lab Chip 2018, 18, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Dalziel, M.; Crispin, M.; Scanlan, C.N.; Zitzmann, N.; Dwek, R.A. Emerging principles for the therapeutic exploitation of glycosylation. Science 2014, 343, 1235681. [Google Scholar] [CrossRef] [PubMed]
- Salanti, A.; Clausen, T.M.; Agerbaek, M.O.; Al Nakouzi, N.; Dahlback, M.; Oo, H.Z.; Lee, S.; Gustavsson, T.; Rich, J.R.; Hedberg, B.J.; et al. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell 2015, 28, 500–514. [Google Scholar] [CrossRef] [PubMed]
- Ernst, B.; Magnani, J.L. From carbohydrate leads to glycomimetic drugs. Nat. Rev. Drug Discov. 2009, 8, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Hudak, J.E.; Bertozzi, C.R. Glycotherapy: New advances inspire a reemergence of glycans in medicine. Chem. Biol. 2014, 21, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Salatino, M.; Girotti, M.R.; Rabinovich, G.A. Glycans pave the way for immunotherapy in triple-negative breast cancer. Cancer Cell 2018, 33, 155–157. [Google Scholar] [CrossRef]
- Bull, C.; Boltje, T.J.; van Dinther, E.A.; Peters, T.; de Graaf, A.M.; Leusen, J.H.; Kreutz, M.; Figdor, C.G.; den Brok, M.H.; Adema, G.J. Targeted delivery of a sialic acid-blocking glycomimetic to cancer cells inhibits metastatic spread. ACS Nano 2015, 9, 733–745. [Google Scholar] [CrossRef]


| Glycan | Structure | Link to Prostate Cancer | References |
|---|---|---|---|
| Example of N-glycan on PSA | 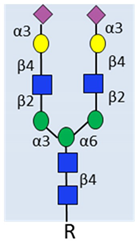 α2-3 sialylated N-glycan | Complex biantennary glycoforms with α2,3-sialic acid have been closely linked to aggressive disease in multiple studies. | [33,34,35]. |
| Sialyl Lewis X (SLeX) |  | Upregulated and linked to poor prognosis in patients. Detected on PSA, MUC1 and PAP in malignant tissue | [49,50,51,52,57]. |
| Sialyl Tn (sTn) |  | Expressed in high-grade prostate tumours. Can reduce prostate cancer cell adhesion. | [58,59,60,61] |
| Core Fucosylation |  | Increased in patient serum. Linked to aggressive disease. | [43,66,67,68] |
| Levels correlate with Fucosyltransferase 8 (FUT8). | [69] | ||
| O-GlcNAcylation |  | Upregulated and linked to poor prognosis in primary prostate cancer. | [70,71] |
| Downregulated in castrate resistant prostate cancer(CRPC). | [72] | ||
| Branched N-glycans |  | Linked to metastasis and disease-free survival. | [73] |
| Predictive biomarker for CRPC. | [74] | ||
| Cryptic N-glycans |  | N-linked oligomannose 9 (Man9) is increased in high-grade tumours and linked to clinical outcome. | [75] |

| Proteoglycan | Link to Prostate Cancer | References |
|---|---|---|
| Versican | Modulates binding to the extracellular matrix (ECM) and enhances motility. Associated with poor prognosis. | [106,110,111,112] |
| Decorin | Suppresses tumour growth by inhibiting both androgen receptor (AR) and epidermal growth factor (EGF). Decreased in prostate cancer. | [106,113,114] |
| Biglycan | Associated with poor prognosis and PTEN deletion. | [107] |
| Lumican | Lumican in stroma tissue suppresses cancer progression. Potential marker in prostate cancer staging. | [115] |
| Perlecan | Linked to disease progression. Upregulates sonic hedgehog signalling. | [116,117] |
| Syndecan-1 | Role in the epithelial-to-mesenchymal transition (EMT). Maintains stability of prostate tumour initiating cells. | [118,119] |
| Poor prognosis. | [108,109] |
| Galectin | Link to Prostate Cancer | References |
|---|---|---|
| Galectin-1 | Upregulated during disease progression. Linked to angiogenesis. | [121] |
| Potential therapeutic target in CRPC. | [125,126] | |
| Galectin-3 | Promotes prostate tumour growth and invasion. Potential diagnostic marker. | [127] |
| High in early stages, lost in advanced disease. May predict biochemical recurrence. | [123,124] | |
| Role in bone metastasis niche. | [122] | |
| Galectin-4 | Linked to metastasis and reduced survival. | [128] |
| Part of O-glycosylation-mediated signalling circuit drives metastatic CRPC. | [129] | |
| Galectin-8 | Linked to metastasis. Proposed as prognostic biomarker. | [130] |
| Glycosylation Enzyme | Role in Glycosylation | Link to Prostate Cancer | References |
|---|---|---|---|
| ST6GALNAC1 | Transfers α-2,6 sialic acid to O-linked GalNAc | Regulated by androgens. Upregulated in tumour tissue. Linked to the synthesis of sTn. Reduced cell adhesion | [60,61,142] |
| GCNT1 | Forms core-2-branched O-linked glycans | Increased in aggressive disease. Closely related to extraprostatic extension and lymph node metastasis. Increases tumour growth on orthotopic inoculation into the mouse prostate. | [133,134] |
| Resistance to NK cell immunity. | [52] | ||
| Regulated by androgens | [132] | ||
| Associated with higher levels of core 2 O sLex in PSA, PAP, and MUC1 | [57] | ||
| Linked to F77 antigen. | [97] | ||
| Detected in post-DRE urine. Indicator of extracapsular extension | [139] | ||
| GCNT2 | Forms core-2-branched O-linked glycans | Linked to invasion. Potential role in integrin signalling. | [143] |
| GALNT7 | Initiation of O-glycosylation | Upregulated in malignant PCa as part of a glycosylation gene signature. | [144] |
| Androgen regulated and linked to prostate cancer cell viability. | [132] | ||
| Correlates with AR-V7 in CRPC. | [138] | ||
| C1GALT1 | Generates the common core 1 O-glycan structure | Part of O-glycosylation mediated signalling circuit that drives CRPC and is linked to poor survival. | [129] |
| ST6Gal1 | Addition of sialic acid to galactose-containing N-glycan | Upregulated. Linked to reduced survival and metastasis. | [135] |
| Regulated by androgens. | [132] | ||
| EDEM3 | Mannose trimming of N-glycans | Upregulated in malignant prostate cancer as part of a glycosylation gene signature. | [144] |
| Androgen regulated and linked to prostate cancer cell viability. | [132] | ||
| MGAT5 | Biosynthesis of β1,6 GlcNAc-branched N-glycans | Link to metastasis in mouse models. | [145] |
| UAP1 | Last enzyme in HBP pathway. Produces UDP-GlcNAc | Highly overexpressed (correlates negatively with Gleason score). Linked to increased UDP-GlcNAc. Protects prostate cancer cells from endoplasmic reticulum (ER) stress. Regulated by androgens. | [88] |
| GNPNAT1 | HBP pathway. Produces UDP-GlcNAc | GNPNAT1 is decreased in CRPC. | [72] |
| FUT6 | Fucosylation | Upregulated in distant metastases. Role in prostate cancer metastasis to bone. | [136] |
| FUT8 | Transfers fucose to core-GlcNAc of the N-glycans | Increased in aggressive prostate cancer and linked to poor prognosis. | [77] |
| Increased in CRPC. | [78] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scott, E.; Munkley, J. Glycans as Biomarkers in Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 1389. https://doi.org/10.3390/ijms20061389
Scott E, Munkley J. Glycans as Biomarkers in Prostate Cancer. International Journal of Molecular Sciences. 2019; 20(6):1389. https://doi.org/10.3390/ijms20061389
Chicago/Turabian StyleScott, Emma, and Jennifer Munkley. 2019. "Glycans as Biomarkers in Prostate Cancer" International Journal of Molecular Sciences 20, no. 6: 1389. https://doi.org/10.3390/ijms20061389
APA StyleScott, E., & Munkley, J. (2019). Glycans as Biomarkers in Prostate Cancer. International Journal of Molecular Sciences, 20(6), 1389. https://doi.org/10.3390/ijms20061389