Analysis of Rice Proteins with DLN Repressor Motif/S
Abstract
:1. Introduction
2. Results and Discussion
2.1. Rice Has a Huge Repertoire of DLN Transcriptional Repressors
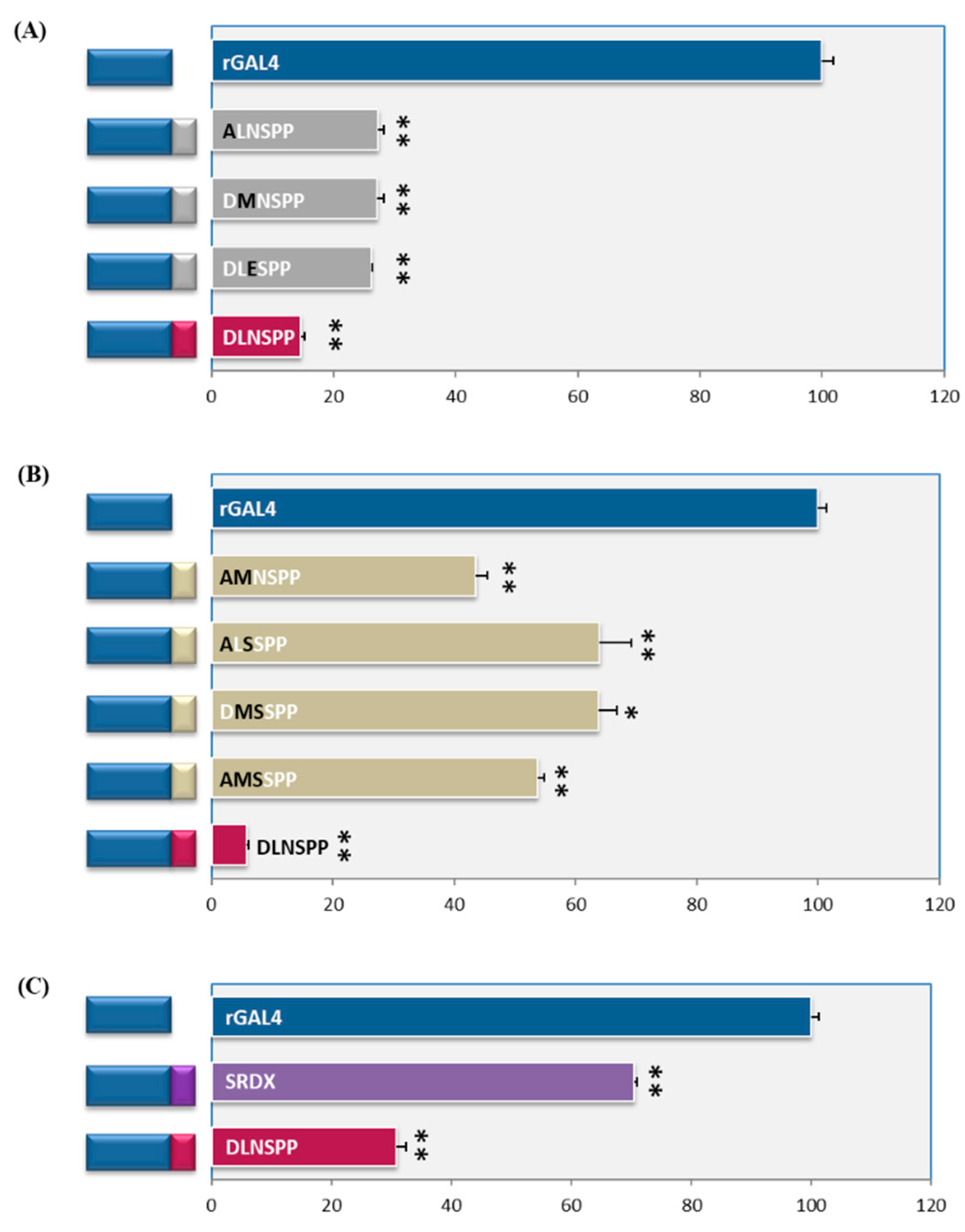
2.2. DLN Motif Is a Strong Repressor
2.3. At Least Two Amino Acids Should Intact for Maximal Repressive Activity
2.4. There Are Subtle Differences amongst DLN Motifs from Rice and Arabidopsis
3. Materials and Methods
3.1. Data Mining
3.2. Construction of Effector Plasmids
3.3. Analysis of Repressive Activity of the Constructs
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Lietz, M.; Hohl, M. How mammalian transcriptional repressors work. Eur. J. Biochem. 2004, 271, 2855–2862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagale, S.; Links, M.G.; Rozwadowski, K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 2010, 152, 1109–1134. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhang, Y.; Han, J.; Luo, J.; Li, G.; Huang, J.; Wu, H.; Tian, Q.; Zhu, Q.; Chen, Y.; et al. The intronic cis element SE1 recruits trans-acting repressor complexes to repress the expression of ELONGATED UPPERMOST INTERNODE1 in rice. Mol. Plant 2018, 11, 720–735. [Google Scholar] [CrossRef]
- Liu, P.; Liu, J.; Dong, H.; Sun, J. Functional regulation of Q by microRNA172 and transcriptional co-repressor TOPLESS in controlling bread wheat spikelet density. Plant Biotechnol. J. 2018, 16, 495–506. [Google Scholar] [CrossRef]
- Chang, G.; Yang, W.; Zhang, Q.; Huang, J.; Yang, Y.; Hu, X. ABI5-BINDING PROTEIN 2 coordinates CONSTANS to delay flowering by recruiting the transcriptional corepressor TPR2. Plant Physiol. 2018. [Google Scholar] [CrossRef]
- Wu, R.; Citovsky, V. Adaptor proteins GIR1 and GIR2. II. Interaction with the co-repressor TOPLESS and promotion of histone deacetylation of target chromatin. Biochem. Biophys. Res. Commun. 2017, 488, 609–613. [Google Scholar] [CrossRef]
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Zhang, T.; Yang, J.; Cubas, P.; Qin, G. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007296. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Sun, Z.; Ding, M.; Logacheva, M.D.; Kreft, I.; Wang, D.; Yan, M.; Shao, J.; Tang, Y.; Wu, Y.; et al. FtSAD2 and FtJAZ1 regulate activity of the FtMYB11 transcription repressor of the phenylpropanoid pathway in Fagopyrum tataricum. New Phytol. 2017, 216, 814–828. [Google Scholar] [CrossRef]
- Ohno, C.K.; Reddy, G.V.; Heisler, M.G.; Meyerowitz, E.M. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 2004, 131, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Moreau, F.; Thevenon, E.; Blanvillain, R.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Dumas, R.; Parcy, F.; Morel, P.; Trehin, C.; Carles, C.C. The Myb-domain protein ULTRAPETALA1 INTERACTING FACTOR 1 controls floral meristem activities in Arabidopsis. Development 2016, 143, 1108–1119. [Google Scholar] [CrossRef]
- Wei, B.; Zhang, J.; Pang, C.; Yu, H.; Guo, D.; Jiang, H.; Ding, M.; Chen, Z.; Tao, Q.; Gu, H.; et al. The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 2015, 25, 121–134. [Google Scholar] [CrossRef]
- Borg, M.; Rutley, N.; Kagale, S.; Hamamura, Y.; Gherghinoiu, M.; Kumar, S.; Sari, U.; Esparza-Franco, M.A.; Sakamoto, W.; Rozwadowski, K.; et al. An EAR-dependent regulatory module promotes male germ cell division and sperm fertility in Arabidopsis. Plant Cell 2014, 26, 2098–2113. [Google Scholar] [CrossRef]
- Kazan, K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 2006, 11, 109–112. [Google Scholar] [CrossRef]
- Maruyama, Y.; Yamoto, N.; Suzuki, Y.; Chiba, Y.; Yamazaki, K.; Sato, T.; Yamaguchi, J. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci. 2013, 213, 79–87. [Google Scholar] [CrossRef]
- Joo, J.; Choi, H.J.; Lee, Y.H.; Kim, Y.K.; Song, S.I. A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta 2013, 238, 155–170. [Google Scholar] [CrossRef]
- Lawrence, S.D.; Novak, N.G.; Jones, R.W.; Farrar, R.R., Jr.; Blackburn, M.B. Herbivory responsive C2H2 zinc finger transcription factor protein StZFP2 from potato. Plant Physiol. Biochem. 2014, 80, 226–233. [Google Scholar] [CrossRef]
- Cao, F.Y.; DeFalco, T.A.; Moeder, W.; Li, B.; Gong, Y.; Liu, X.M.; Taniguchi, M.; Lumba, S.; Toh, S.; Shan, L.; et al. Arabidopsis ETHYLENE RESPONSE FACTOR 8 (ERF8) has dual functions in ABA signaling and immunity. BMC Plant Biol. 2018, 18, 211. [Google Scholar] [CrossRef]
- Sun, Y.W.; Tee, C.S.; Ma, Y.H.; Wang, G.; Yao, X.M.; Ye, J. Attenuation of histone methyltransferase KRYPTONITE-mediated transcriptional gene silencing by Geminivirus. Sci. Rep. 2015, 5, 16476. [Google Scholar] [CrossRef]
- Thireault, C.; Shyu, C.; Yoshida, Y.; St Aubin, B.; Campos, M.L.; Howe, G.A. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant J. 2015, 82, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Quan, R.; Pan, X.; Wan, L.; Huang, R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta 2013, 237, 1443–1451. [Google Scholar] [CrossRef]
- Paponov, I.A.; Teale, W.; Lang, D.; Paponov, M.; Reski, R.; Rensing, S.A.; Palme, K. The evolution of nuclear auxin signalling. BMC Evol. Biol. 2009, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Ryu, H.; Hwang, I.; Wang, Z.Y. TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat. Commun. 2014, 5, 4140. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, S.F.; Parish, R.W. Regulation of gene expression by manipulating transcriptional repressor activity using a novel CoSRI technology. Plant Biotechnol. J. 2017, 15, 879–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiratsu, K.; Matsui, K.; Koyama, T.; Ohme-Takagi, M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003, 34, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Hommel, M.; Khalil-Ahmad, Q.; Jaimes-Miranda, F.; Mila, I.; Pouzet, C.; Latché, A.; Pech, J.C.; Bouzayen, M.; Regad, F. Over-expression of a chimeric gene of the transcriptional co-activator MBF1 fused to the EAR repressor motif causes developmental alteration in Arabidopsis and tomato. Plant Sci. 2008, 175, 168–177. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Li, P.F.; Chen, M.K.; Lee, Y.I.; Yang, C.H. FOREVER YOUNG FLOWER negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol. 2015, 168, 1666–1683. [Google Scholar] [CrossRef] [PubMed]
- Hur, Y.S.; Um, J.H.; Kim, S.; Kim, K.; Park, H.J.; Lim, J.S.; Kim, W.Y.; Jun, S.E.; Yoon, E.K.; Lim, J.; et al. Arabidopsis thaliana homeobox 12 (ATHB12), a homeodomain-leucine zipper protein, regulates leaf growth by promoting cell expansion and endoreduplication. New Phytol. 2015, 205, 316–328. [Google Scholar] [CrossRef]
- Agarwal, P.; Arora, R.; Ray, S.; Singh, A.K.; Singh, V.P.; Takatsuji, H.; Kapoor, S.; Tyagi, A.K. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 2007, 65, 467–485. [Google Scholar] [CrossRef] [PubMed]
- Mathew, I.E.; Agarwal, P. May the fittest protein evolve: Favoring the plant-specific origin and expansion of NAC transcription factors. Bioessays 2018, 40, e1800018. [Google Scholar] [CrossRef] [PubMed]
- Mathew, I.E.; Das, S.; Mahto, A.; Agarwal, P. Three rice NAC transcription factors heteromerize and are associated with seed size. Front. Plant Sci. 2016, 7, 1638. [Google Scholar] [CrossRef]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takagi, M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Hiratsu, K.; Mitsuda, N.; Matsui, K.; Ohme-Takagi, M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 2004, 321, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Kapoor, S.; Tyagi, A.K. Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 2011, 33, 189–202. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2510. [Google Scholar] [CrossRef]
- Danino, Y.M.; Even, D.; Ideses, D.; Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta 2015, 1849, 1116–1131. [Google Scholar] [CrossRef] [PubMed]
- Duttke, S.H.C. Evolution and diversification of the basal transcription machinery. Trends Biochem. Sci. 2015, 40, 127–129. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, S.; Liu, S.; Yu, M.; Su, H.; Shu, H.; Li, X. Global analysis of ankyrin repeat domain C3HC4-type RING finger gene family in plants. PLoS ONE 2013, 8, e58003. [Google Scholar] [CrossRef]
- Chaharbakhshi, E.; Jemc, J.C. Broad-complex, tramtrack, and bric-à-brac (BTB) proteins: Critical regulators of development. Genesis 2016, 54, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Rozwadowski, K. Small yet effective: The ethylene responsive element binding factor-associated amphiphilic repression (EAR) motif. Plant Signal. Behav. 2010, 5, 691–694. [Google Scholar] [CrossRef] [PubMed]
- Daware, A.; Das, S.; Srivastava, R.; Badoni, S.; Singh, A.K.; Agarwal, P.; Parida, S.K.; Tyagi, A.K. An efficient strategy combining SSR markers- and advanced QTL-seq-driven QTL mapping unravels candidate genes regulating grain weight in rice. Front. Plant Sci. 2016, 7, 1535. [Google Scholar] [CrossRef]
- Malik, N.; Dwivedi, N.; Singh, A.K.; Parida, S.K.; Agarwal, P.; Thakur, J.K.; Tyagi, A.K. An integrated genomic strategy delineates candidate mediator genes regulating grain size and weight in rice. Sci. Rep. 2016, 6, 23253. [Google Scholar] [CrossRef]
- Sharma, R.; Agarwal, P.; Ray, S.; Deveshwar, P.; Sharma, P.; Sharma, N.; Nijhawan, A.; Jain, M.; Singh, A.K.; Singh, V.P.; et al. Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct. Integr. Genomics 2012, 12, 229–248. [Google Scholar] [CrossRef] [PubMed]
- Mahto, A.; Mathew, I.E.; Agarwal, P. Decoding the Transcriptome of Rice Seed During Development; InTech: London, UK, 2017; pp. 26–44. [Google Scholar]
- Agarwal, P.; Parida, S.K.; Mahto, A.; Das, S.; Mathew, I.E.; Malik, N.; Tyagi, A.K. Expanding frontiers in plant transcriptomics in aid of functional genomics and molecular breeding. Biotechnol. J. 2014, 9, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Parida, S.K.; Raghuvanshi, S.; Kapoor, S.; Khurana, P.; Khurana, J.P.; Tyagi, A.K. Rice improvement through genome-based functional analysis and molecular breeding in India. Rice (NY) 2016, 9, 1. [Google Scholar] [CrossRef]
- Hanna-Rose, W.; Hansen, U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996, 12, 229–234. [Google Scholar] [CrossRef]
- Nakano, T.; Fujisawa, M.; Shima, Y.; Ito, Y. The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 2014, 65, 3111–3119. [Google Scholar] [CrossRef] [Green Version]
- Amalraj, A.; Luang, S.; Kumar, M.Y.; Sornaraj, P.; Eini, O.; Kovalchuk, N.; Bazanova, N.; Li, Y.; Yang, N.; Eliby, S.; et al. Change of function of the wheat stress-responsive transcriptional repressor TaRAP2.1L by repressor motif modification. Plant Biotechnol. J. 2016, 14, 820–832. [Google Scholar] [CrossRef]
- Sakamoto, H.; Maruyama, K.; Sakuma, Y.; Meshi, T.; Iwabuchi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004, 136, 2734–2746. [Google Scholar] [CrossRef]
- Ogata, T.; Kida, Y.; Tochigi, M.; Matsushita, Y. Analysis of the cell death-inducing ability of the ethylene response factors in group VIII of the AP2/ERF family. Plant Sci. 2013, 209, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Szemenyei, H.; Hannon, M.; Long, J.A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 2008, 319, 1384–1386. [Google Scholar] [CrossRef] [PubMed]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Jeong, N.; Suh, S.J.; Kim, M.H.; Lee, S.; Moon, J.K.; Kim, H.S.; Jeong, S.C. Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell 2012, 24, 4807–4818. [Google Scholar] [CrossRef]
- Ciftci-Yilmaz, S.; Morsy, M.R.; Song, L.; Coutu, A.; Krizek, B.A.; Lewis, M.W.; Warren, D.; Cushman, J.; Connolly, E.L.; Mittler, R. The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 2007, 282, 9260–9268. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Hiratsu, K.; Todaka, D.; Nakashima, K.; Yamaguchi-Shinozaki, K.; Ohme-Takagi, M. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 2006, 4, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Yasuhara, M.; Geekiyanage, S.; Ogura, Y.; Kiyosue, T. Overexpression of the chimeric gene of the floral regulator CONSTANS and the EAR motif repressor causes late flowering in Arabidopsis. Plant Cell Rep. 2007, 26, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Gao, G.; Zhong, Y.; Guo, A.; Zhu, Q.; Tang, W.; Zheng, W.; Gu, X.; Wei, L.; Luo, J. DRTF: A database of rice transcription factors. Bioinformatics 2006, 22, 1286–1287. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.; Mathew, I.E.; Verma, A.; Tyagi, A.K.; Agarwal, P. Analysis of Rice Proteins with DLN Repressor Motif/S. Int. J. Mol. Sci. 2019, 20, 1600. https://doi.org/10.3390/ijms20071600
Singh P, Mathew IE, Verma A, Tyagi AK, Agarwal P. Analysis of Rice Proteins with DLN Repressor Motif/S. International Journal of Molecular Sciences. 2019; 20(7):1600. https://doi.org/10.3390/ijms20071600
Chicago/Turabian StyleSingh, Purnima, Iny Elizebeth Mathew, Ankit Verma, Akhilesh K. Tyagi, and Pinky Agarwal. 2019. "Analysis of Rice Proteins with DLN Repressor Motif/S" International Journal of Molecular Sciences 20, no. 7: 1600. https://doi.org/10.3390/ijms20071600
APA StyleSingh, P., Mathew, I. E., Verma, A., Tyagi, A. K., & Agarwal, P. (2019). Analysis of Rice Proteins with DLN Repressor Motif/S. International Journal of Molecular Sciences, 20(7), 1600. https://doi.org/10.3390/ijms20071600




