Structural Variations in LysM Domains of LysM-RLK PsK1 May Result in a Different Effect on Pea–Rhizobial Symbiosis Development
Abstract
:1. Introduction
2. Results
2.1. Effect of R. Leguminosarum bv. Viciae Strain A34 and Its Derivatives nodE (A42), nodO (A67) and nodE nodO (A91) Rhizobial Strains on Pea Mutants Impaired in the k1 Gene
2.2. Analysis of Dimerization between PsSYM10 and PsK1 Carrying Replacements in LysM1, LysM3 and Kinase Domains in Various Systems
2.2.1. Transient Co-Production of Pea LysM-RLKs in N. Benthamiana Leaves
2.2.2. Yeast Two-Hybrid Assay (GAL4 Transcription Factor-Based Assay)
2.3. Effect of LysM-RLK PsK1 on Symbiosis Development with AM Fungus
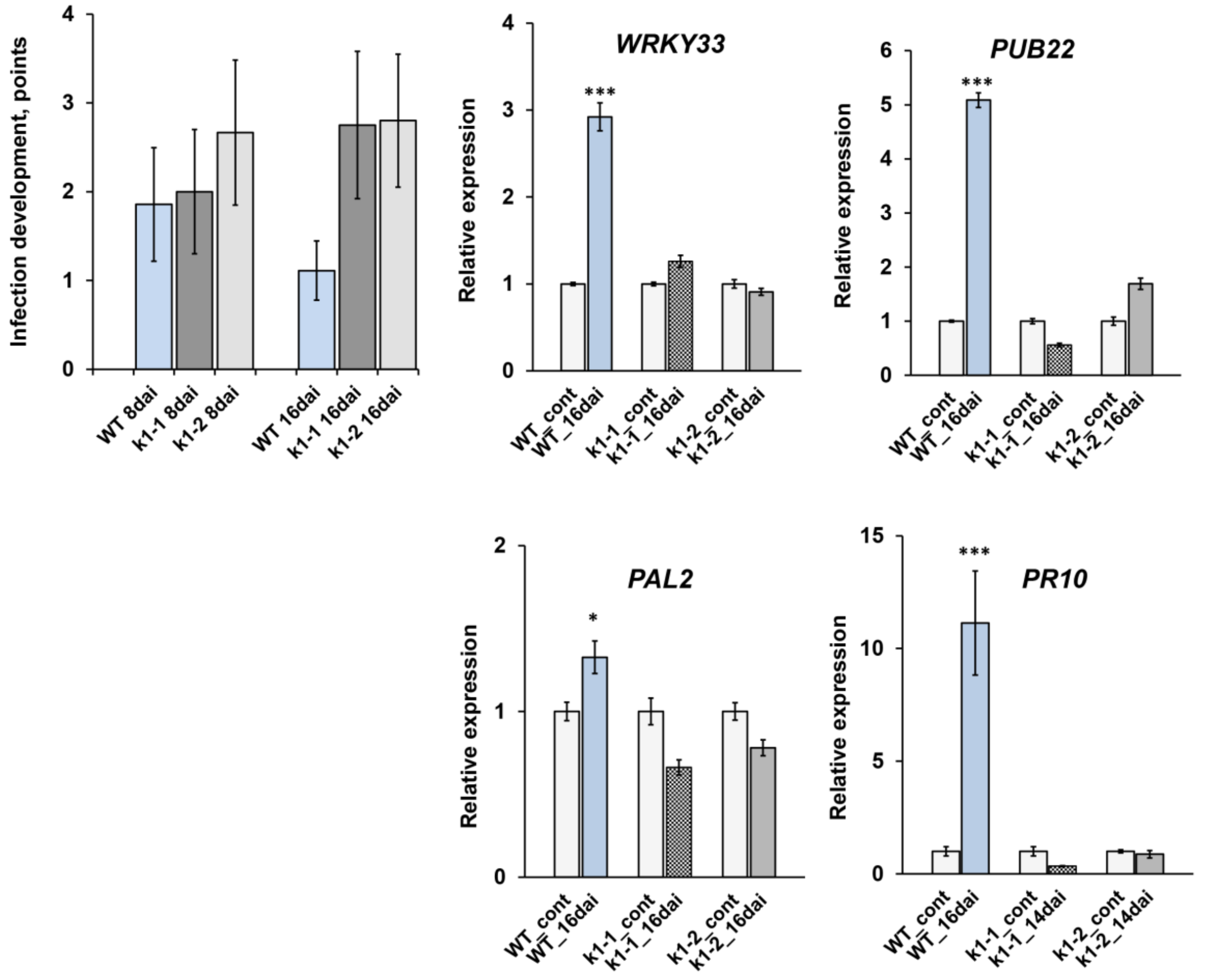
2.4. LysM-RLK PsK1 May Be Involved in Pea Resistance to Fungal Pathogen Fusarium Culmorum 334
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Inoculation
4.2. Plant Material and Growth Conditions
4.3. RNA Extraction and Quantitative Reverse Transcription PCR (qRT-PCR)
4.4. Generation of Constructs for Plant and Yeast Transformation
4.5. Transient Protein Expression in Nicotiana Benthamiana Leaves
4.6. Western Blot Analysis of Leaf Tissues
4.7. Yeast Two-Hybrid Assay (GAL4 Transcription Factor-Based Assay)
4.8. Histochemical Staining and Microscopy
4.9. Analysis of Mycorrhizal Colonization
4.10. Fusarium Culmorum Infection
4.11. Statistical Methods and Computer Software
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miya, A.; Albert, P.; Shinya, T.; Desaki, Y.; Ichimura, K.; Shirasu, K.; Narusaka, Y.; Kawakami, N.; Kaku, H.; Shibuya, N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 19613–19618. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, X.-C.; Neece, D.; Ramonell, K.M.; Clough, S.; Kim, S.-Y.; Stacey, M.G.; Stacey, G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 2008, 20, 471–481. [Google Scholar] [CrossRef]
- Kaku, H.; Nishizawa, Y.; Ishii-Minami, N.; Akimoto-Tomiyama, C.; Dohmae, N.; Takio, K.; Minami, E.; Shibuya, N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 11086–11091. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, G.; Wang, J.; Jin, C.; Chang, J.; et al. Chitin-induced dimerization activates a plant immune receptor. Science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liang, Y.; Tanaka, K.; Nguyen, C.T.; Jedrzejczak, R.P.; Joachimiak, A.; Stacey, G. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife 2014. [Google Scholar] [CrossRef]
- Miyata, K.; Kozaki, T.; Kouzai, Y.; Ozawa, K.; Ishii, K.; Asamizu, E.; Okabe, Y.; Umehara, Y.; Miyamoto, A.; Kobae, Y.; et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014, 55, 1864–1872. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.-F.; Ao, Y.; Qu, J.; Li, Z.; Su, J.; Zhang, Y.; Liu, J.; Feng, D.; Qi, K.; et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 2012, 24, 3406–3419. [Google Scholar] [CrossRef]
- Willmann, R.; Lajunen, H.M.; Erbs, G.; Newman, M.-A.; Kolb, D.; Tsuda, K.; Katagiri, F.; Fliegmann, J.; Bono, J.-J.; Cullimore, J.V.; et al. Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 2011, 108, 19824–19829. [Google Scholar] [CrossRef]
- Hayafune, M.; Berisio, R.; Marchetti, R.; Silipo, A.; Kayama, M.; Desaki, Y.; Arima, S.; Squeglia, F.; Ruggiero, A.; Tokuyasu, K.; et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA 2014, 111, E404–E413. [Google Scholar] [CrossRef]
- Liu, S.; Wang, J.; Han, Z.; Gong, X.; Zhang, H.; Chai, J. Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure 2016, 24, 1192–1200. [Google Scholar] [CrossRef]
- Leppyanen, I.V.; Shakhnazarova, V.Y.; Shtark, O.Y.; Vishnevskaya, N.A.; Tikhonovich, I.A.; Dolgikh, E.A. Receptor-like kinase LYK9 in Pisum sativum L. is the CERK1-like receptor that controls both plant immunity and AM symbiosis development. Int. J. Mol. Sci. 2017, 19, 8. [Google Scholar] [CrossRef]
- Bozsoki, Z.; Cheng, J.; Feng, F.; Gysel, K.; Vinther, M.; Andersen, K.R.; Oldroyd, G.; Blaise, M.; Radutoiu, S.; Stougaard, J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. USA 2017, 114, E8118–E8127. [Google Scholar] [CrossRef]
- Zipfel, C.; Oldroyd, G.E.D. Plant signalling in symbiosis and immunity. Nature 2017, 543, 328–336. [Google Scholar] [CrossRef]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef]
- Broghammer, A.; Krusell, L.; Blaise, M.; Sauer, J.; Sullivan, J.T.; Maolanon, N.; Vinther, M.; Lorentzen, A.; Madsen, E.B.; Jensen, K.J.; et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 2012, 109, 13859–13864. [Google Scholar] [CrossRef]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.D.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Dénarié, J.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003, 34, 495–506. [Google Scholar] [CrossRef]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef]
- Arrighi, J.-F. The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006, 142, 265–279. [Google Scholar] [CrossRef]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, V.; Radutoiu, S.; Madsen, L.H.; Rychagova, T.; Ovchinnikova, E.; Borisov, A.; Tikhonovich, I.; Stougaard, J. The Pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development. Mol. Plant Microbe Interact. 2008, 21, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Fliegmann, J.; Canova, S.; Lachaud, C.; Uhlenbroich, S.; Gasciolli, V.; Pichereaux, C.; Rossignol, M.; Rosenberg, C.; Cumener, M.; Pitorre, D.; et al. Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chem. Biol. 2013, 8, 1900–1906. [Google Scholar] [CrossRef]
- Malkov, N.; Fliegmann, J.; Rosenberg, C.; Gasciolli, V.; Timmers, A.C.J.; Nurisso, A.; Cullimore, J.; Bono, J.-J. Molecular basis of lipo-chitooligosaccharide recognition by the lysin motif receptor-like kinase LYR3 in legumes. Biochem. J. 2016, 473, 1369–1378. [Google Scholar] [CrossRef]
- Kirienko, A.N.; Porozov, Y.B.; Malkov, N.V.; Akhtemova, G.A.; Le Signor, C.; Thompson, R.; Saffray, C.; Dalmais, M.; Bendahmane, A.; Tikhonovich, I.A.; et al. Role of a receptor-like kinase K1 in pea rhizobium symbiosis development. Planta 2018, 248, 1101–1120. [Google Scholar] [CrossRef]
- Ardourel, M. Rhizobium meliloti Lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 1994, 6, 1357–1374. [Google Scholar] [CrossRef]
- Walker, S.A.; Downie, J.A. Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol. Plant Microbe Interact. 2000, 13, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Geurts, R.; Fedorova, E.; Bisseling, T. Nod factor signaling genes and their function in the early stages of rhizobium infection. Curr. Opin. Plant Biol. 2005, 8, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Bensmihen, S.; de Billy, F.; Gough, C. Contribution of NFP LysM domains to the recognition of nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS ONE 2011, 6, e26114. [Google Scholar] [CrossRef] [PubMed]
- Pietraszewska-Bogiel, A.; Lefebvre, B.; Koini, M.A.; Klaus-Heisen, D.; Takken, F.L.W.; Geurts, R.; Cullimore, J.V.; Gadella, T.W.J. Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS ONE 2013, 8, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Moling, S.; Pietraszewska-Bogiel, A.; Postma, M.; Fedorova, E.; Hink, M.A.; Limpens, E.; Gadella, T.W.J.; Bisseling, T. Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 2014, 26, 4188–4199. [Google Scholar] [CrossRef]
- Fliegmann, J.; Jauneau, A.; Pichereaux, C.; Rosenberg, C.; Gasciolli, V.; Timmers, A.C.J.; Burlet-Schiltz, O.; Cullimore, J.; Bono, J.-J. LYR3, a high-affinity LCO-binding protein of Medicago truncatula, interacts with LYK3, a key symbiotic receptor. FEBS Lett. 2016, 590, 1477–1487. [Google Scholar] [CrossRef] [PubMed]
- Mulder, L.; Lefebvre, B.; Cullimore, J.; Imberty, A. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 2006, 16, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.H.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin–oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Igolkina, A.A.; Porozov, Y.B.; Chizhevskaya, E.P.; Andronov, E.E. Structural insight into the role of mutual polymorphism and conservatism in the contact zone of the NFR5–K1 heterodimer with the nod factor. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Dalmais, M.; Schmidt, J.; Le Signor, C.; Moussy, F.; Burstin, J.; Savois, V.; Aubert, G.; Brunaud, V.; de Oliveira, Y.; Guichard, C.; et al. UTILLdb, a Pisum sativum in silico forward and reverse genetics tool. Genome Biol. 2008, 9, R43. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, W.; Sun, J.; Feng, F.; Deng, Y.; He, Z.; Oldroyd, G.E.D.; Wang, E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015, 81, 258–267. [Google Scholar] [CrossRef]
- Sun, J.; Miller, J.B.; Granqvist, E.; Wiley-Kalil, A.; Gobbato, E.; Maillet, F.; Cottaz, S.; Samain, E.; Venkateshwaran, M.; Fort, S.; et al. Activation of symbiosis signaling by Arbuscular Mycorrhizal fungi in legumes and rice. Plant Cell 2015, 27, 823–838. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef]
- Czaja, L.F.; Hogekamp, C.; Lamm, P.; Maillet, F.; Martinez, E.A.; Samain, E.; Denarie, J.; Kuster, H.; Hohnjec, N. Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by Arbuscular Mycorrhizal Fungal Lipochitooligosaccharides. Plant Physiol. 2012, 159, 1671–1685. [Google Scholar] [CrossRef]
- Rey, T.; Nars, A.; Bonhomme, M.; Bottin, A.; Huguet, S.; Balzergue, S.; Jardinaud, M.-F.; Bono, J.-J.; Cullimore, J.; Dumas, B.; et al. NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 2013, 198, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Downie, J.A.; Knight, C.D.; Johnston, A.W.B.; Rossen, L. Identification of genes and gene products involved in the nodulation of peas by Rhizobium leguminosarum. MGG Mol. Gen. Genet. 1985, 198, 255–262. [Google Scholar] [CrossRef]
- Economou, A.; Davies, A.E.; Johnston, A.W.B.; Downie, J.A. The Rhizobium leguminosarum biovar viciae nodO gene can enable a nodE mutant of Rhizobium leguminosarum biovar trifolii to nodulate vetch. Microbiology 1994, 140, 2341–2347. [Google Scholar] [CrossRef]
- Schneider, A.; Walker, S.A.; Poyser, S.; Sagan, M.; Ellis, T.H.N.; Downie, J.A. Genetic mapping and functional analysis of a nodulation-defective mutant (sym19) of pea (Pisum sativum L.). Mol. Gen. Genet. MGG 1999, 262, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Knox, M.R.; Edwards, A.; Hogg, B.; Ellis, T.H.N.; Wei, G.; Downie, J.A. Natural variation in host-specific nodulation of pea is associated with a haplotype of the SYM37 LysM-type receptor-like kinase. Mol. Plant Microbe Interact. 2011, 24, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Sutton, J.M.; Lea, E.J.; Downie, J.A. The nodulation-signaling protein NodO from Rhizobium leguminosarum biovar viciae forms ion channels in membranes. Proc. Natl. Acad. Sci. USA 1994, 91, 9990–9994. [Google Scholar] [CrossRef] [PubMed]
- Knight, C.D.; Rossen, L.; Robertson, J.G.; Wells, B.; Downie, J.A. Nodulation inhibition by Rhizobium leguminosarum multicopy nodABC genes and analysis of early stages of plant infection. J. Bacteriol. 1986. [Google Scholar] [CrossRef]
- Downie, J.A.; Surin, B.P. Either of two nod gene loci can complement the nodulation defect of a nod deletion mutant of Rhizobium leguminosarum bv viciae. Mol. Gen. Genet. 1990, 222, 81–86. [Google Scholar] [CrossRef]
- Shaw, S.L. Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 2003, 131, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Spaink, H.P.; Sheeley, D.M.; van Brussel, A.A.N.; Glushka, J.; York, W.S.; Tak, T.; Geiger, O.; Kennedy, E.P.; Reinhold, V.N.; Lugtenberg, B.J.J. A novel highly unsaturated fatty acid moiety of lipo-oligosaccharide signals determines host specificity of Rhizobium. Nature 1991, 354, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.B.; Antolín-Llovera, M.; Grossmann, C.; Ye, J.; Vieweg, S.; Broghammer, A.; Krusell, L.; Radutoiu, S.; Jensen, O.N.; Stougaard, J.; et al. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 2011, 65, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Halter, T.; Imkampe, J.; Mazzotta, S.; Wierzba, M.; Postel, S.; Bücherl, C.; Kiefer, C.; Stahl, M.; Chinchilla, D.; Wang, X.; et al. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 2014, 24, 134–143. [Google Scholar] [CrossRef]
- Leong, S.A.; Williams, P.H.; Ditta, G.S. Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res. 1985, 13, 5965–5976. [Google Scholar] [CrossRef] [PubMed]
- Van Brussel, A.A.N.; Planque, K.; Quispel, A. The wall of Rhizobium leguminosarum in bacteroid and free-living forms. J. Gen. Microbiol. 1977, 101, 51–56. [Google Scholar] [CrossRef]
- Van Brussel, A.A.; Tak, T.; Wetselaar, A.; Pees, E.; Wijffelman, C. Small leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Sci. Lett. 1982, 27, 317–325. [Google Scholar] [CrossRef]
- Dolgikh, E.A.; Shaposhnikov, A.I.; Dolgikh, A.V.; Gribchenko, E.S.; Bodyagina, K.B.; Yuzhikhin, O.S.; Tikhonovich, I.A. Identification of Pisum sativum L. cytokinin and auxin metabolic and signaling genes, and an analysis of their role in symbiotic nodule development. Int. J. Plant Physiol. Biochem. 2017, 9, 22–35. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Boivin, C.; Camut, S.; Malpica, C.A.; Truchet, G.; Rosenberg, C. Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 1990, 2, 1157–1170. [Google Scholar] [CrossRef]
- Shtark, O.Y.; Sulima, A.S.; Zhernakov, A.I.; Kliukova, M.S.; Fedorina, J.V.; Pinaev, A.G.; Kryukov, A.A.; Akhtemova, G.A.; Tikhonovich, I.A.; Zhukov, V.A. Arbuscular mycorrhiza development in pea (Pisum sativum L.) mutants impaired in five early nodulation genes including putative orthologs of NSP1 and NSP2. Symbiosis 2016, 68, 129–144. [Google Scholar] [CrossRef]
- Trouvelot, A.; Kough, J..; Gianinazzi-Pearson, V. Mesure du taux de mycorhization VA d’un systeme radiculaire. Recherche de methodes d’estimation ayant une significantion fonctionnelle. Mycorrhizae Physiol. Genet. 1986, 39, 418–420. [Google Scholar] [CrossRef]
- Vierheilig, H.; Coughlan, A.P.; Wyss, U.; Piche, Y. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 1998, 64, 5004–5007. [Google Scholar] [PubMed]
- Park, H.-J.; Floss, D.S.; Levesque-Tremblay, V.; Bravo, A.; Harrison, M.J. Hyphal branching during arbuscule development requires RAM1. Plant Physiol. 2015, 169, 2774–2788. [Google Scholar] [CrossRef] [PubMed]








| Mutant Line | Mutation | DNA Position | Protein Position | Localization |
|---|---|---|---|---|
| 885 (k1-1) | G → A | 1445 | G332D | Kinase domain |
| 817 (k1-2) | C → T | 571 | P169S | LysM3 motif of ECD |
| 2265 (k1-3) | C → T | 242 | S59F | LysM1 motif of ECD |
| Variant | Root-Hair Deformations and Curling | |||
|---|---|---|---|---|
| A34 (wild type) | A42 (nodE) | A67 (nodO) | A91 (nodE nodO) | |
| Cameor (wild type) | 292.2 ± 2.6 b | 426.6 ± 3.5 a | 297.65 ± 4.9 b | 124.94 ± 3.3 d |
| k1-1 mutant | 4.49 ± 0.2 f | 5.84 ± 0.27 f | 0 | 0 |
| k1-2 mutant | 122.7 ± 2.7 d | 188.7 ± 3.3 c | 37.03 ± 1.33 e | 30.81 ± 0.7 * e |
| k1-3 mutant | 297.2 ± 2.6 b | 434.1 ± 5.8 a | 294.9 ± 1.5 b | 120.2 ± 2 d |
| Infection Thread Growth (number of ITs Aborted in Epidermis and Outer Cortex is Enclosed in Brackets) | ||||
| A34 (wild type) | A42 (nodE) | A67 (nodO) | A91 (nodE nodO) | |
| Cameor (wild type) | 22.9 ± 0.9 fg | 25.6 ± 0.9 f | 16.7 ± 0.5 g | 1028.99 ± 6.09 a (1020.82 ± 8.89)*** |
| k1-1 mutant** | 2.85 ± 0.2 h | 4.28 ± 0.4 h | 0 | 0 |
| k1-2 mutant | 141.2 ± 2.1 b | 139.1± 2.65 b | 33.3 ± 1.6 d | 0 |
| (99.6 ± 1.51) | (103.2±1.5) | (31.65 ± 1.4) | ||
| k1-3 mutant | 43.9 ± 1.25 ce | 46.54± 1.1 c | 34.7 ± 1.6 de | 1035.6 ± 3.1 a (1029.8 ± 2.8)*** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirienko, A.N.; Vishnevskaya, N.A.; Kitaeva, A.B.; Shtark, O.Y.; Kozyulina, P.Y.; Thompson, R.; Dalmais, M.; Bendahmane, A.; Tikhonovich, I.A.; Dolgikh, E.A. Structural Variations in LysM Domains of LysM-RLK PsK1 May Result in a Different Effect on Pea–Rhizobial Symbiosis Development. Int. J. Mol. Sci. 2019, 20, 1624. https://doi.org/10.3390/ijms20071624
Kirienko AN, Vishnevskaya NA, Kitaeva AB, Shtark OY, Kozyulina PY, Thompson R, Dalmais M, Bendahmane A, Tikhonovich IA, Dolgikh EA. Structural Variations in LysM Domains of LysM-RLK PsK1 May Result in a Different Effect on Pea–Rhizobial Symbiosis Development. International Journal of Molecular Sciences. 2019; 20(7):1624. https://doi.org/10.3390/ijms20071624
Chicago/Turabian StyleKirienko, Anna N., Nadezhda A. Vishnevskaya, Anna B. Kitaeva, Oksana Yu. Shtark, Polina Yu. Kozyulina, Richard Thompson, Marion Dalmais, Abdelhafid Bendahmane, Igor A. Tikhonovich, and Elena A. Dolgikh. 2019. "Structural Variations in LysM Domains of LysM-RLK PsK1 May Result in a Different Effect on Pea–Rhizobial Symbiosis Development" International Journal of Molecular Sciences 20, no. 7: 1624. https://doi.org/10.3390/ijms20071624
APA StyleKirienko, A. N., Vishnevskaya, N. A., Kitaeva, A. B., Shtark, O. Y., Kozyulina, P. Y., Thompson, R., Dalmais, M., Bendahmane, A., Tikhonovich, I. A., & Dolgikh, E. A. (2019). Structural Variations in LysM Domains of LysM-RLK PsK1 May Result in a Different Effect on Pea–Rhizobial Symbiosis Development. International Journal of Molecular Sciences, 20(7), 1624. https://doi.org/10.3390/ijms20071624







