1. Introduction
Upper urinary tract urothelial carcinoma (UTUC) is a relatively rare disease compared with urinary bladder urothelial carcinoma. Advanced UTUC is often associated with poor oncologic outcome [
1]. Chemotherapy is the current standard therapy for advanced UTUC in the neoadjuvant or adjuvant setting [
2]. However, the high prevalence of renal insufficiency in UTUC is a clinical challenge [
3]. For those patients with renal insufficiency, which means they are ineligible for chemotherapy, radiation therapy is sometimes an adjunct strategy. Further exploration of treatment targets sensitive to treatment multimodality may help improve clinical outcome.
The secreted protein acidic rich in cysteine-like 1 (SPARCL1) protein belongs to the SPARC-related family of matricellular proteins. The human
SPARCL1 gene was initially discovered in high endothelial venules from tonsils [
4]. Several studies have described the role of SPARCL1 in the prognosis of colorectal, gastric, ovarian, and prostate cancers [
5,
6,
7,
8]. The function of SPARCL1 is not completely known, but studies have suggested that it may modulate high endothelial cell adhesion to the basement membrane as an antiadhesive protein [
9]. Additionally, SPARCL1 was reported to inhibit the progression of cells from G
1 to S phase and to negatively regulate cell proliferation [
10]. It is also a tumour suppressor as it induces cell differentiation possibly via MET, which represses the aggressiveness of CRCs [
5].
To the best of our best knowledge, no studies have yet reported the role of SPARCL1 in urothelial carcinoma. We found that high-stage/high-grade UTUC samples had significant SPARCL1 hypermethylation compared with normal urothelium adjacent to low-stage/low-grade specimens. The objective of this study was to identify the role of SPARCL1 in advanced UTUC.
3. Discussion
Advanced UTUC is often associated with early local regional recurrence or distant metastasis, leading to poor survival outcomes even after standard radical nephroureterectomy [
1]. Therefore, the treatment strategy needs to include perioperative chemotherapy to improve survival [
2]. However, perioperative renal insufficiency, especially after radical surgery, was observed in patients with UTUC. Such a high prevalence of renal function impairment leads to an inadequate dose of cisplatin-based chemotherapy [
3]. Though an immune checkpoint inhibitor was developed as an alternative treatment choice for patients with urothelial carcinoma ineligible for cisplatin therapy, the response rate was still limited [
14]. Urothelial carcinoma is not always curable if distant metastasis develops. Moreover, urinary bladder cancer recurrence is not uncommon after UTUC management, owing to the multifocal characteristics of urothelial carcinoma [
15]. Potential advanced bladder cancer development often causes major disability in these patients after radical surgery [
16]. Further investigation into tumorigenesis is needed to improve cancer control and prevent further metastasis of UTUC.
DNA methylation is the C5 methylation of cytosine bases in a CpG dinucleotide. DNA methylation serves as an epigenetic mark to organize the complex human genome. In cancers, DNA methylation patterns are generally disturbed compared with the corresponding normal tissues. DNA hypermethylation of CpG island promoters is commonly associated with transcriptional repression of the affected promoter, leading to decreased gene expression or alternative promoter use [
17]. Therefore, the hypermethylation of tumour suppressor genes may be associated with tumorigenesis due to epigenetic change. Such methods have been investigated mostly in urinary bladder urothelial carcinoma by studying tumour or urine samples to serve as prognostic biomarkers or diagnostic tools for early detection [
18]. The application of DNA methylation in UTUC is less frequently reported, owing to its relatively rare incidence. In Taiwan, an unusually high prevalence of UTUC has been reported, and it is a major public health problem [
19]. The study of DNA methylation with adequate validation cohorts may help in preventing or detecting UTUC.
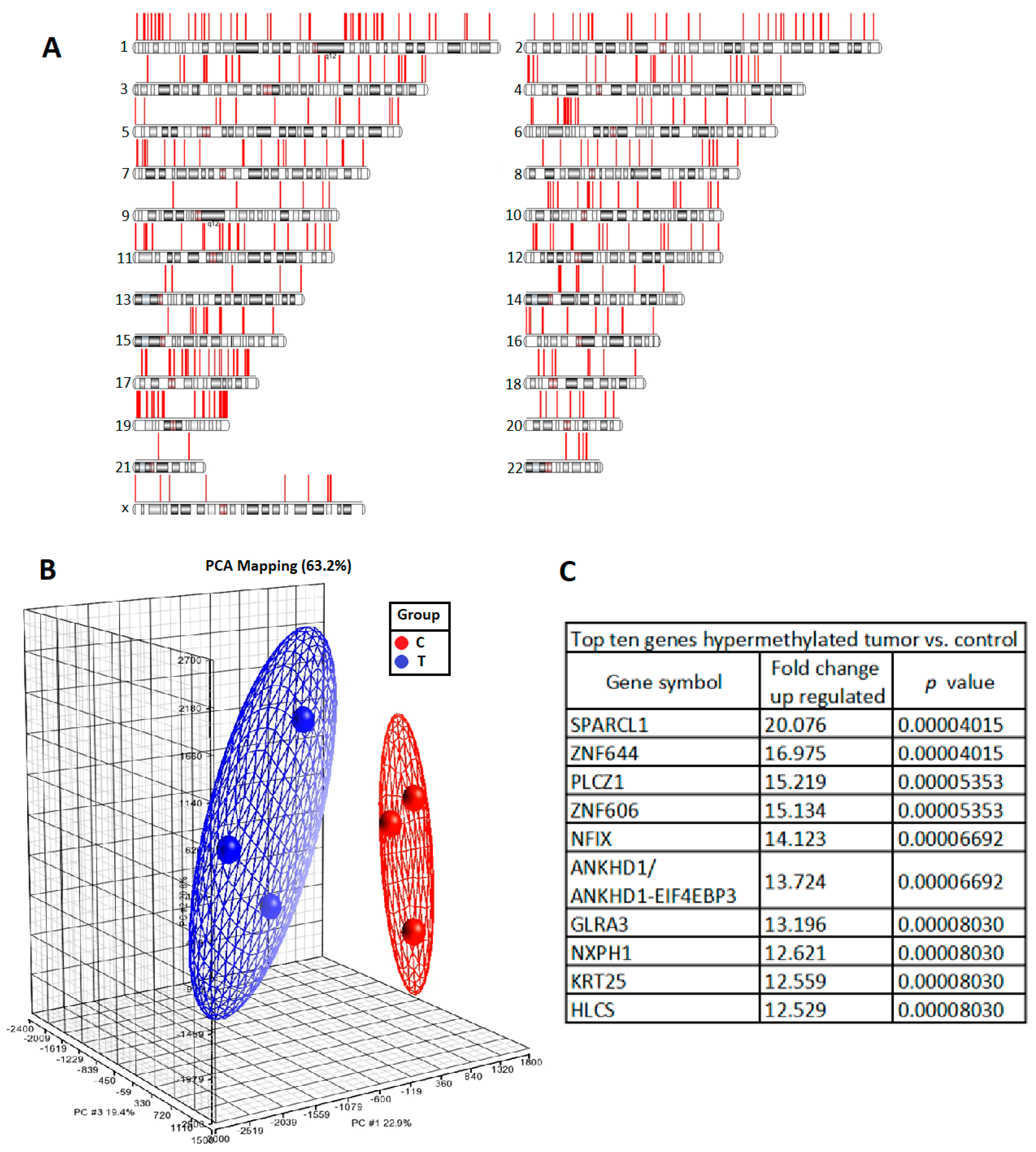
To identify any epigenetic change of the tumorigenesis of UTUC, we selected three tumours of high-grade and high-stage UTUC and three normal urothelial tissue samples adjacent to low-grade and low-stage UTUC by comparing the methylation intensities of promoter regions. The methylation of
SPARCL1 was significantly greater in UTUC than in normal urothelium. Pyrosequencing-based analysis of our prospectively collected UTUC and normal urothelial tissue samples further proved significant
SPARCL1 hypermethylation in UTUC. Such a high prevalence of
SPARCL1 hypermethylation in the UTUC samples indicates that the loss of SPARCL1 function might be considered to have clinical utility in the assessment of UTUC behavior. In this study, the 5-year recurrence-free survival of advanced UTUC was approximately 60% in the SPARCL1-positive group, whereas it was approximately 35% in the SPARCL1-negative group (presented as Kaplan Meier plot in
Figure 3D). Both SPARCL1 presentation and tumour architecture were significant in regard to systemic UTUC recurrence in our institutional cohort.
The prognostic value of SPARCL1 and tumour architecture is independent in multivariate analysis, compared to other currently known prognostic factors. The BFTC909 cell line originates from an aggressive renal pelvic tumour with low SPARCL1 expression in Taiwan. Our in vitro study shows that its aggressive behaviour can be reversed by SPARCL1 overexpression in colony formation assays and migration assays. A lower N-cadherin/E-cadherin ratio, which indicates less epithelial–mesenchymal transition and cell migration, is also compatible with less malignant behaviour after SPARCL1 overexpression. The sensitivity to radiation and chemotherapy can be also observed in SPARCL1 overexpressing cells. Further correlation studies between SPARCL1 expression and treatment response for locally advanced UTUC might provide more information in the future. Therefore, from the preliminary result, SPARCL1 could be considered a prognostic biomarker for advanced UTUC and further identification of the outcome of the clinical benefit of chemotherapy or radiation on patients with positive SPARCL1 UTUC needs further clinical trial for validation of its prognostic utility.
The role of SPARCL1 is commonly discussed in gastrointestinal malignancies. A meta-analysis of eight studies including a total of 2356 patients revealed that its expression is associated with less lymph node and distant metastasis. The negative association between the SPARCL1 expression and poor tumour differentiation indicates the major role of aggressive tumour behavior [
20]. In addition, the prognostic impact of SPARCL1 expression in ovarian and prostate cancers has also been reported [
7,
8]. The mechanism of SPARCL1 in cancer behaviour in these studies involved cell growth, proliferation, differentiation, cell cycle inhibition, and regulation of the microenvironment [
9,
10,
11]. We examined the status of SPARCL1 methylation status of several UC cell lines and found that SPARCL1 methylation was more common in aggressive UC cell lines. However, to the best of our knowledge, no report on the prognostic role of SPARCL1 in urothelial carcinoma has been published to date. On the basis of the prospective tissue analysis and retrospective cohort validation in this study, we believe that the prognostic role of SPARCL1 expression should not be overlooked in UTUC.
This is the first study on the impact of SPARCL1 expression on advanced UTUC. In this study, we demonstrated that SPARCL1 may have clinical utility as a prognostic biomarker that is independently associated with UTUC recurrence. The major limitation is its retrospective single-institution design with limited case numbers and only a Taiwanese population represented. However, we selected a retrospective advanced UTUC cohort in this study with a primary focus on pT3 disease, instead of pT4 disease, because these patients can often be treated with surgical resection for representative pathologic review. Unlike low-stage disease, pT3 UTUC often recurs within 2 years, and the average follow-up duration in this study is long enough to observe the oncologic outcome. The prognostic effect of biomarkers in low-stage UTUC may interfere with the curative effect radical surgery. In addition, we carefully excluded perioperative chemotherapy in order to observe natural UTUC behaviour. The endpoint of this study was systemic disease recurrence instead of patient survival because palliative or salvage therapy after systemic disease recurrence might add more confounding factors to the analysis of the prognostic role of biomarkers. In addition, the methylation status of cell lines in this study are not all UTUC. To better understand the methylation status between the aggressive and non-aggressive cell lines, the methylation status in primary culture cells should be validated.
The significant correlation between SPARCL1 expression and advanced UTUC behaviour is an interesting finding. In this study, careful selection of a retrospective advanced UTUC cohort, and in vitro cancer behaviour analysis proved the important role of SPARCL1 in UTUC. The conclusion about the oncologic impact of SPARCL1 is similar in many other cancer studies. Studies on the prevalence of SPARCL1 expression in UTUC in patients of other races are still needed to clarify the role of application in clinical practice worldwide. In Taiwan, SPARCL1 methylation is commonly found in UTUC. Further therapeutic agents modulating SPARCL1 methylation or development of SPARCL1-based screening may help with UTUC treatment or prevention.
4. Materials and Methods
4.1. Study Patient Selection
First, we started to collect the prospective UTUC samples since 2016. We included 6 patients for DNA methylation analysis. Then 25 patients for DNA methylation validation (pyrosequencing) and 55 patients using real-time PCR detected SPARCL1 are from prospectively collected samples (
Supplementary File 2). The relationship between SPARCL1 expression and pathological stage/distant metastasis in retrospective whole stage distribution TMA cohort (
Supplementary File 2). However, for clinical translation, we want to identify the clinical prognostic role of SPARCL1 for advanced UTUC. Therefore, the IHC information from 78 patients for clinical advanced UTUC outcome cohort is from the full representative slide by pathologist’s review. The 78 patients with solitary ureteral or renal pelvic pT3N0M0 urothelial carcinomas without perioperative neoadjuvant or adjuvant therapy were included in the study. Pathological features and clinical outcome assessment were as previously described [
21]. This study was approved by Kaohsiung Chang Gung Medical Center Institutional Review Board since 01/Mar/2016 (approval number 106-4117C). Informed consent was obtained preoperatively.
4.2. DNA Methylation Analysis
DNA from the clinical tissues was isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). After passing the quality control criteria, the DNA samples were first subjected to immunoprecipitation using proteins with a methyl-CpG-binding domain. Then, the enriched DNA fragments were amplified through polymerase chain reaction (PCR) and loaded onto the GeneChip Human Promoter 1.0R tiling array (Affymetrix, Santa Clara, CA, USA). We determined the difference in methylation between tumour samples of high-grade and high-stage UTUC (n = 3) and normal urothelium adjacent to low-grade and low-stage UTUC (n = 3) by comparing the probe intensities of promoter regions.
4.3. Pyrosequencing-Based Bisulfite PCR Analysis
A total of 500 ng of DNA from each sample was treated using the EZ DNA Methylation-Lightning Kits Bisulfite Conversion System (Zymo Research, Irvine, CA, USA) and the converted DNA was eluted in 20 μL of the elution buffer. Bisulfite conversion was performed in the dark at 98 °C for 10 min and 64 °C for 3.5 h, followed by desulphonation of the converted DNA. Gene amplification was performed using the HotStarTaq® Master Mix Kit (Qiagen). The CpG1, CpG2, and CpG3 sites of SPARCL1 pyrosequencing primers used were as follows: forward: 5′-GGTGTGTGGGAAAAGTTTTAGAT-3′; reverse: 5′-CCAAATTTCCAATTTCTCTTAAACC-3′; sequencing: 5′-TTATTTAATTTTTTTGAGTTTTAT-3′. The CpG4 of SPARCL1 pyrosequencing primers used were as follows: forward: 5′-GTGGGAAAAGTTTTAGATTTAGAGTT -3′; reverse: 5′-CCAAATTTCCAATTTCTCTTAAACC-3′; sequencing: 5′-AAAAGTGAGATAGATTAAGTATA-3′. Amplification conditions were as follows: 95 °C for 15 min, 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, 45 cycles. To assay DNA methylation levels of the SPARCL1 promoter, bisulfite sequencing was performed on the PyroMark Q24 instrument (Qiagen). Relative levels of methylation at each CpG site were analyzed with PyroMark Q24 version 2.0.6 software.
4.4. Immunohistochemistry and Patient Grouping
A human UTUC TMA containing 103 specimens (with triplicate cores for each sample) was provided from Chang Gung Medical Foundation Kaohsiung Chang Gung Memorial Hospital Tissue Bank. Immunostaining for SPARCL1 was performed on a fully automated Bond-Max system (Leica Microsystems, Wetzlar, Germany). Slides carrying tissue sections cut from formalin-fixed, paraffin-embedded tissue microarray blocks were dried for 1 hour at 60 °C. These slides were then covered by Bond Universal Covertiles and placed into the Bond-Max instrument. All subsequent steps were performed automatically by the instrument according to the manufacturer’s instructions (Leica Microsystems) according to a previous report [
21]. The following procedure was used: (1) Deparaffinization of tissue on slides by rinsing with Bond Dewax Solution at 72 °C; (2) heat-induced epitope retrieval (antigen unmasking) with Bond Epitope Retrieval Solution 2 for 10 min at 100 °C; (3) peroxide block placement on the slides for 10 min at room temperature; (4) incubation with a mouse monoclonal anti-SPARCL-1 antibody at a dilution of 1:100 for 60 min at room temperature; (5) incubation with Post Primary reagent for 10 min at room temperature; (6) Bond Polymer placement on the slides for 8 min at room temperature; (7) color development with 3,3′-diaminobenzidine tetrahydrochloride (DAB) as a chromogen for 3 min at room temperature; and (8) hematoxylin counterstaining for 1 minutes. Slides were mounted and examined by light microscopy.
The immunoreactivity scoring was based on the intensity of positive staining using a 3-point scale: 0–10%, 0; 11–50%, 1; 51–80%, 2; and >80%, 3. The identification of SPARCL1 expression was performed by a uropathologist (Dr. Min-Tse Sung) and urooncologist (Dr. Hao Lun Luo) in our institution.
4.5. RNA Isolation and Real-Time PCR
RNA was extracted by means of QIAGEN RNA purification kit from UTUC clinical tissue and cell lines. Five microgram RNA of each sample will be reverse transcribed using RevertAidTM H Minus Reverse Transcriptase (Fermentas, Waltham, MA, USA). Real-time PCR was performed using SYBR Green PCR master mix (Life Technologies, Carlsbad, CA, USA) and ABI 7500 sequence detection system (Life Technologies). The real-time PCR primers: SPARCL1 forward: 5′-GTGAAGGCAACATGAGGGTGCA-3′; SPARCL1 reverse primer: 5′-GTTGGAGGACAAGTCACTGGATC-3′. GAPDH forward: 5′-GTCTCCTCTGACTTCAACAGCG-3′; GAPDH reverse primer: 5′-ACCACCCTGTTGCTGTAGCCAA-3′. All primers were purchased from OriGene (Rockville, MD, USA) and checked for specificity using BLAST (NCBI). Exon/intron junctions were spanned.
4.6. Western Blotting Assays
Cells were lysed directly in a modified RIPA buffer containing a protease inhibitor mixture (Roche, Basel, Switzerland). The relative protein concentration in the supernatants was related to standard BSA concentrations determined by BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). For each lane of 8~10% SDS–PAGE gel, 40 ug protein of cell lysates were loaded, separated and subsequently transferred onto Immobilon-P Transfer Membrane (Millipore, Burlington, MA, USA). The membranes were probed with specific antibodies. The primary antibodies were against SPARCL1 (Abcam, Cambridge, MA, UK), Vimentin (Cell Signaling Technology, Danvers, MA, USA), N-cadherin (Cell Signaling Technology, Danvers, MA, USA), E-cadherin (Cell Signaling Technology, Danvers, MA, USA), β-actin (Chemicon, Temecula, CA) and GAPDH (GeneTex, Irvine, CA, USA). The primary antibodies were used: SPARCL1 (1:500), Vimentin (1:1000), N-cadherin (1:1000), E-cadherin (1:1000), β-actin (1:5000) and GAPDH (1:5000). The secondary antibodies were added and incubated for 2 h and then visualized using chemiluminescence. Enhanced chemiluminescence (ECL) western blotting reagents were obtained from Pierce Biotechnology (Rockford, IL, USA).
4.7. Cell Line Study
SV-HUC-1, RT4, J82, BFTC909, and T24 cell lines were purchased from Bioresource Collection and Research Center (BCRC). The plasmid expressing shSPARCL1 and the cDNAs coding SPARCL1 wild-type were purchased from OriGene. All constructs were verified by DNA sequencing analyses. Plasmids were isolated by QIAGEN Plasmid Mini Kit and transfection was performed by using TurboFect transfection reagents (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The migration assay we used has been described in detail previously [
21]. SV-HUC-1, RT4, and BFTC909 cells were treated with vehicle or 2, 5, 10 μM 5-Aza-2′-deoxycytidine (Sigma-Aldrich, Saint Louis, MO, USA) for 2 days. Media with vehicle or 5-Aza-2′-deoxycytidine was changed daily. The histology of UTUC is urothelial carcinoma. The included cell lines RT4/J82/T24 are bladder urothelial carcinomas. However, we can only obtain the aggressive UTUC cell line (BFTC 909) from Taiwan instead of non-aggressive UTUC cell line (actually all the available UTUC cell lines in the world are aggressive in behavior, which is compatible with the finding from clinical practice). Therefore, we examined other UC cell lines to identify the SPARCL methylation in the aggressive UC cell line (BFTC909/T24/J82) and relatively non-aggressive UC cell line (RT4).
4.8. Colony Formation Assay
Cells of different conditions (Control or SPARCL1 knockdown/overexpression). The BFTC909 cells were seeded in the 25T flasks. For the clonogenic assay, 0, 2, 4, 6, and 8 Gy were delivered using 6MV photon beam from the linear accelerator. They were irradiated from one posterior-anterior portal with 1cm bolus. We seeded different cell number (50–7200) according to the radiation doses in six-well plates. After 13 days, we used methanol to fix cells and 0.5% crystal violet to stain colonies and used distilled water wash twice. Cells were counted using a stereomicroscope.
4.9. Statistical Analysis
SPSS Statistics version 17 software (SPSS, Chicago, IL, USA) was used for all statistical analyses. The chi-squared test and independent samples
t-test were used for inter-group comparisons. The Kaplan–Meier method with the log-rank test was used for time-to-event analysis. Multivariate Cox regression analysis was used to assess the independent roles of perioperative factors in systemic recurrence [
21]. A
p value less than or equal to 0.05 was used to define a statistically significant result. The observed endpoint in this study was systemic recurrence before initiation of systemic anticancer therapy.