Laccases with Variable Properties from Different Strains of Steccherinum ochraceum: Does Glycosylation Matter?
Abstract
:1. Introduction
2. Results
2.1. Purification and Identification
2.2. Comparison of Physicochemical and Catalytic Properties
2.3. Identification of Gene Sequences
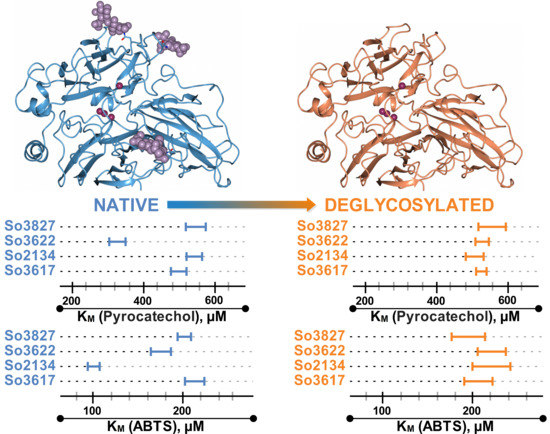
2.4. Assessment of Influence of Glycosylation
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 2,6-DMP | 2,6-dimetoxyphenol |
| ABTS | 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) |
| GlcNAc | N-acetylglucoseamine |
| Man | Mannose |
| PDB | Protein Data Bank |
| SNP | Single Nucleotide Polymorphism |
References
- Messerschmidt, A. Blue copper oxidases. Adv. Inorg. Chem. 1993, 40, 121–185. [Google Scholar]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper oxidases and oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar] [CrossRef]
- Riva, S. Laccases: Blue enzymes for green chemistry. Trends Biotechnol. 2006, 24, 219–226. [Google Scholar] [CrossRef]
- Rivera-Hoyos, C.M.; Morales-Álvarez, E.D.; Poutou-Piñales, R.A.; Pedroza-Rodríguez, A.M.; RodrÍguez-Vázquez, R.; Delgado-Boada, J.M. Fungal laccases. Fungal Biol. Rev. 2013, 27, 67–82. [Google Scholar] [CrossRef]
- Giardina, P.; Faraco, V.; Pezzella, C.; Piscitelli, A.; Vanhulle, S.; Sannia, G. Laccases: A never-ending story. Cell. Mol. Life Sci. 2010, 67, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Brijwani, K.; Rigdon, A.; Vadlani, P.V. Fungal laccases: Production, function, and applications in food processing. Enzym. Res. 2010, 2010, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Alcalde, M. Laccase: A multi-purpose biocatalyst at the forefront of biotechnology. Microb. Biotechnol. 2017, 10, 1457–1467. [Google Scholar] [CrossRef] [PubMed]
- Pezzella, C.; Guarino, L.; Piscitelli, A. How to enjoy laccases. Cell. Mol. Life Sci. 2015, 72, 923–940. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, W.; Ng, T.B.; Deng, X.; Lin, J.; Ye, X. Laccases: Production, expression regulation, and applications in pharmaceutical biodegradation. Front. Microbiol. 2017, 8, 832. [Google Scholar] [CrossRef] [PubMed]
- Mogharabi, M.; Faramarzi, M.A. Laccase and laccase-mediated systems in the synthesis of organic compounds. Adv. Synth. Catal. 2014, 356, 897–927. [Google Scholar] [CrossRef]
- Viswanath, B.; Rajesh, B.; Janardhan, A.; Kumar, A.P.; Narasimha, G. Fungal laccases and their applications in bioremediation. Enzym. Res. 2014, 2014, 163242. [Google Scholar] [CrossRef] [PubMed]
- Fillat, Ú.; Ibarra, D.; Eugenio, M.; Moreno, A.; Tomás-Pejó, E.; Martín-Sampedro, R. Laccases as a potential tool for the efficient conversion of lignocellulosic biomass: A review. Fermentation 2017, 3, 17. [Google Scholar] [CrossRef]
- Kunamneni, A.; Plou, F.; Ballesteros, A.; Alcalde, M. Laccases and their applications: A patent review. Recent Pat. Biotechnol. 2008, 2, 10–24. [Google Scholar] [CrossRef]
- Yashas, S.R.; Shivakumara, B.P.; Udayashankara, T.H.; Krishna, B.M. Laccase biosensor: Green technique for quantification of phenols in wastewater. Orient. J. Chem. 2018, 34, 631–637. [Google Scholar]
- Rodríguez Couto, S.; Toca Herrera, J.L. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Senthivelan, T.; Kanagaraj, J.; Panda, R.C. Recent trends in fungal laccase for various industrial applications: An eco-friendly approach—A review. Biotechnol. Bioprocess Eng. 2016, 21, 19–38. [Google Scholar] [CrossRef]
- Upadhyay, P.; Shrivastava, R.; Agrawal, P.K. Bioprospecting and biotechnological applications of fungal laccase. 3 Biotech 2016, 6. [Google Scholar] [CrossRef]
- Courty, P.E.; Hoegger, P.J.; Kilaru, S.; Kohler, A.; Buée, M.; Garbaye, J.; Martin, F.; Kües, U. Phylogenetic analysis, genomic organization, and expression analysis of multi-copper oxidases in the ectomycorrhizal basidiomycete Laccaria bicolor. New Phytol. 2009, 182, 736–750. [Google Scholar] [CrossRef]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct subfamilies. Curr. Genet. 2006, 50, 45–60. [Google Scholar] [CrossRef]
- Uzan, E.; Nousiainen, P.; Balland, V.; Sipila, J.; Piumi, F.; Navarro, D.; Asther, M.; Record, E.; Lomascolo, A. High redox potential laccases from the ligninolytic fungi Pycnoporus coccineus and Pycnoporus sanguineus suitable for white biotechnology: From gene cloning to enzyme characterization and applications. J. Appl. Microbiol. 2010, 108, 2199–2213. [Google Scholar]
- Tinoco, R.; Pickard, M.A. Kinetic differences of purified laccases from six Pleurotus ostreatus strains. Lett. Appl. Microbiol. 2001, 32, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Glazunova, O.A.; Shakhova, N.V.; Psurtseva, N.V.; Moiseenko, K.V.; Kleimenov, S.Y.; Fedorova, T.V. White-rot basidiomycetes Junghuhnia nitida and Steccherinum bourdotii: Oxidative potential and laccase properties in comparison with Trametes hirsuta and Coriolopsis caperata. PLoS ONE 2018, 13, e0197667. [Google Scholar] [CrossRef] [PubMed]
- Glazunova, O.A.; Polyakov, K.M.; Moiseenko, K.V.; Kurzeev, S.A.; Fedorova, T.V. Structure-function study of two new middle-redox potential laccases from basidiomycetes Antrodiella faginea and Steccherinum murashkinskyi. Int. J. Biol. Macromol. 2018, 118, 406–418. [Google Scholar] [CrossRef]
- Myasoedova, N.M.; Chernykh, A.M.; Psurtseva, N.V.; Belova, N.V.; Golovleva, L.A. New efficient producers of fungal laccases. Appl. Biochem. Microbiol. 2008, 44, 73–77. [Google Scholar] [CrossRef]
- Chernykh, A.M.; Myasoedova, N.M.; Kolomytseva, M.; Ferraroni, M.; Briganti, F.; Scozzafava, A.; Golovleva, L. Laccase isoforms with unusual properties from the basidiomycete Steccherinum ochraceum strain 1833. J. Appl. Microbiol. 2008, 105, 2065–2075. [Google Scholar] [CrossRef]
- Glazunova, O.; Trushkin, N.; Moiseenko, K.; Filimonov, I.; Fedorova, T. Catalytic efficiency of basidiomycete laccases: Redox potential versus substrate-binding pocket structure. Catalysts 2018, 8, 152. [Google Scholar] [CrossRef]
- Osipov, E.; Polyakov, K.; Kittl, R.; Shleev, S.; Dorovatovsky, P.; Tikhonova, T.; Hann, S.; Ludwig, R.; Popov, V. Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2913–2923. [Google Scholar] [CrossRef] [PubMed]
- Mate, D.M.; Alcalde, M. Laccase engineering: From rational design to directed evolution. Biotechnol. Adv. 2015, 33, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Kogi, H.; Tsujimura, S.; Sakurai, T. Modifications of laccase activities of copper efflux oxidase, CueO by synergistic mutations in the first and second coordination spheres of the type I copper center. Biochem. Biophys. Res. Commun. 2013, 431, 393–397. [Google Scholar] [CrossRef]
- Vite-Vallejo, O.; Palomares, L.A.; Dantán-González, E.; Ayala-Castro, H.G.; Martínez-Anaya, C.; Valderrama, B.; Folch-Mallol, J. The role of N-glycosylation on the enzymatic activity of a Pycnoporus sanguineus laccase. Enzym. Microb. Technol. 2009, 45, 233–239. [Google Scholar] [CrossRef]
- Maestre-Reyna, M.; Liu, W.C.; Jeng, W.Y.; Lee, C.C.; Hsu, C.A.; Wen, T.N.; Wang, A.H.J.; Shyur, L.F. Structural and functional roles of glycosylation in fungal laccase from Lentinus sp. PLoS ONE 2015, 10, 1–28. [Google Scholar] [CrossRef]
- Glazunova, O.A.; Polyakov, K.M.; Fedorova, T.V.; Dorovatovskii, P.V.; Koroleva, O.V. Elucidation of the crystal structure of Coriolopsis caperata laccase: Restoration of the structure and activity of the native enzyme from the T2-depleted form by copper ions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 854–861. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef]



| Laccase | Kinetic Parameters | ABTS | Pyrocatechol | Guaiacol | 2,6-DMP | Syringaldazine |
|---|---|---|---|---|---|---|
| So3120 | KM, µM | 175 ± 23 | 497 ± 31 | 2665 ± 216 | 17 ± 3 | 5.4 ± 0.8 |
| kcat, s−1 | 325 ± 28 | 406 ± 24 | 165 ± 20 | 110 ± 14 | 105 ± 12 | |
| kcat/KM, s−1·mM−1 | 1857 ± 290 | 817 ± 70 | 62 ± 9 | 6471 ± 1273 | 19,444 ± 3617 | |
| So3174 | KM, µM | 81 ± 9 | 576 ± 86 | 2230 ± 205 | 20 ± 2 | 3.8 ± 0.6 |
| kcat, s−1 | 240 ± 21 | 373 ± 27 | 152 ± 13 | 101 ± 9 | 104 ± 11 | |
| kcat/KM, s−1·mM−1 | 2963 ± 416 | 648 ± 108 | 68 ± 9 | 5050 ± 755 | 27,368 ± 5091 | |
| So2134 | KM, µM | 100 ± 6 | 540 ± 22 | 2060 ± 168 | 21 ± 2 | 3.7 ± 0.5 |
| kcat, s−1 | 254 ± 23 | 400 ± 35 | 221 ± 20 | 166 ± 14 | 91 ± 7 | |
| kcat/KM, s−1·mM−1 | 2540 ± 276 | 741 ± 72 | 107 ± 13 | 7905 ± 1034 | 24,595 ± 3929 | |
| So3827 | KM, µM | 200 ± 7 | 545 ± 26 | 3565 ± 363 | 24 ± 3 | 6.7 ± 0.9 |
| kcat, s−1 | 276 ± 31 | 394 ± 37 | 237 ± 22 | 165 ± 15 | 113 ± 12 | |
| kcat/KM, s−1·mM−1 | 1380 ± 162 | 723 ± 76 | 66 ± 9 | 6875 ± 981 | 16,866 ± 2872 | |
| So3398 | KM, µM | 209 ± 27 | 495 ± 41 | 2426 ± 225 | 17 ± 3 | 5.6 ± 0.8 |
| kcat, s−1 | 281 ± 26 | 450 ± 40 | 262 ± 24 | 152 ± 12 | 119 ± 11 | |
| kcat/KM, s−1·mM−1 | 1344 ± 214 | 909 ± 110 | 108 ± 14 | 8941 ± 1516 | 21,250 ± 3565 | |
| So3617 | KM, µM | 212 ± 10 | 495 ± 21 | 2383 ± 257 | 22 ± 2 | 5.2 ± 0.6 |
| kcat, s−1 | 228 ± 21 | 340 ± 31 | 197 ± 20 | 134 ± 11 | 89 ± 10 | |
| kcat/KM, s−1·mM−1 | 1075 ± 111 | 687 ± 69 | 83 ± 12 | 6091 ± 836 | 17,115 ± 2691 | |
| So3622 | KM, µM | 174 ± 11 | 323 ± 25 | 2717 ± 272 | 23 ± 3 | 5.5 ± 0.7 |
| kcat, s−1 | 252 ± 20 | 365 ± 34 | 218 ± 17 | 140 ± 18 | 83 ± 7 | |
| kcat/KM, s−1·mM−1 | 1448 ± 147 | 1130 ± 137 | 80 ± 10 | 6087 ± 1163 | 15,091 ± 2213 |
| Laccase | N-Glycosylation Sites | ||
|---|---|---|---|
| Asn182 | Asn414 | Asn436 | |
| So3120 N * | (Man)5–8(GlcNAc)2 | (Man)6–8(GlcNAc)2 | (Man)6–8(GlcNAc)2 |
| So3120 D * | GlcNAc | GlcNAc | n.d. ** |
| So3174 N | (Man)5–7(GlcNAc)2 | (Man)6–8(GlcNAc)2 | (Man)5–8(GlcNAc)2 |
| So3174 D | GlcNAc | GlcNAc | n.d. |
| So2134 N | (Man)5–7(GlcNAc)2 | (Man)5–8(GlcNAc)2 | n.d. |
| So2134 D | GlcNAc | GlcNAc | n.d. |
| So3827 N | (Man)5–7(GlcNAc)2 | (Man)6–8(GlcNAc)2 | (Man)7–8(GlcNAc)2 |
| So3827 D | GlcNAc | GlcNAc | GlcNAc |
| So3398 N | (Man)5–7(GlcNAc)2 | (Man)6–8(GlcNAc)2 | (Man)5–7(GlcNAc)2 |
| So3398 D | GlcNAc | GlcNAc | n.d. |
| So3617 N | (Man)5–7(GlcNAc)2 | (Man)6–8(GlcNAc)2 | n.d. |
| So3617 D | n.d. | GlcNAc | n.d. |
| So3622 N | GlcNAc, (Man)5–7(GlcNAc)2 | (Man)6–8(GlcNAc)2 | (Man)7–8(GlcNAc)2 |
| So3622 D | n.d. | GlcNAc | n.d. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glazunova, O.A.; Moiseenko, K.V.; Kamenihina, I.A.; Isaykina, T.U.; Yaropolov, A.I.; Fedorova, T.V. Laccases with Variable Properties from Different Strains of Steccherinum ochraceum: Does Glycosylation Matter? Int. J. Mol. Sci. 2019, 20, 2008. https://doi.org/10.3390/ijms20082008
Glazunova OA, Moiseenko KV, Kamenihina IA, Isaykina TU, Yaropolov AI, Fedorova TV. Laccases with Variable Properties from Different Strains of Steccherinum ochraceum: Does Glycosylation Matter? International Journal of Molecular Sciences. 2019; 20(8):2008. https://doi.org/10.3390/ijms20082008
Chicago/Turabian StyleGlazunova, Olga A., Konstantin V. Moiseenko, Inna A. Kamenihina, Tatyana U. Isaykina, Alexander I. Yaropolov, and Tatyana V. Fedorova. 2019. "Laccases with Variable Properties from Different Strains of Steccherinum ochraceum: Does Glycosylation Matter?" International Journal of Molecular Sciences 20, no. 8: 2008. https://doi.org/10.3390/ijms20082008
APA StyleGlazunova, O. A., Moiseenko, K. V., Kamenihina, I. A., Isaykina, T. U., Yaropolov, A. I., & Fedorova, T. V. (2019). Laccases with Variable Properties from Different Strains of Steccherinum ochraceum: Does Glycosylation Matter? International Journal of Molecular Sciences, 20(8), 2008. https://doi.org/10.3390/ijms20082008