Screening for Cholesterol-Lowering Probiotics from Lactic Acid Bacteria Isolated from Corn Silage Based on Three Hypothesized Pathways
Abstract
1. Introduction
2. Results
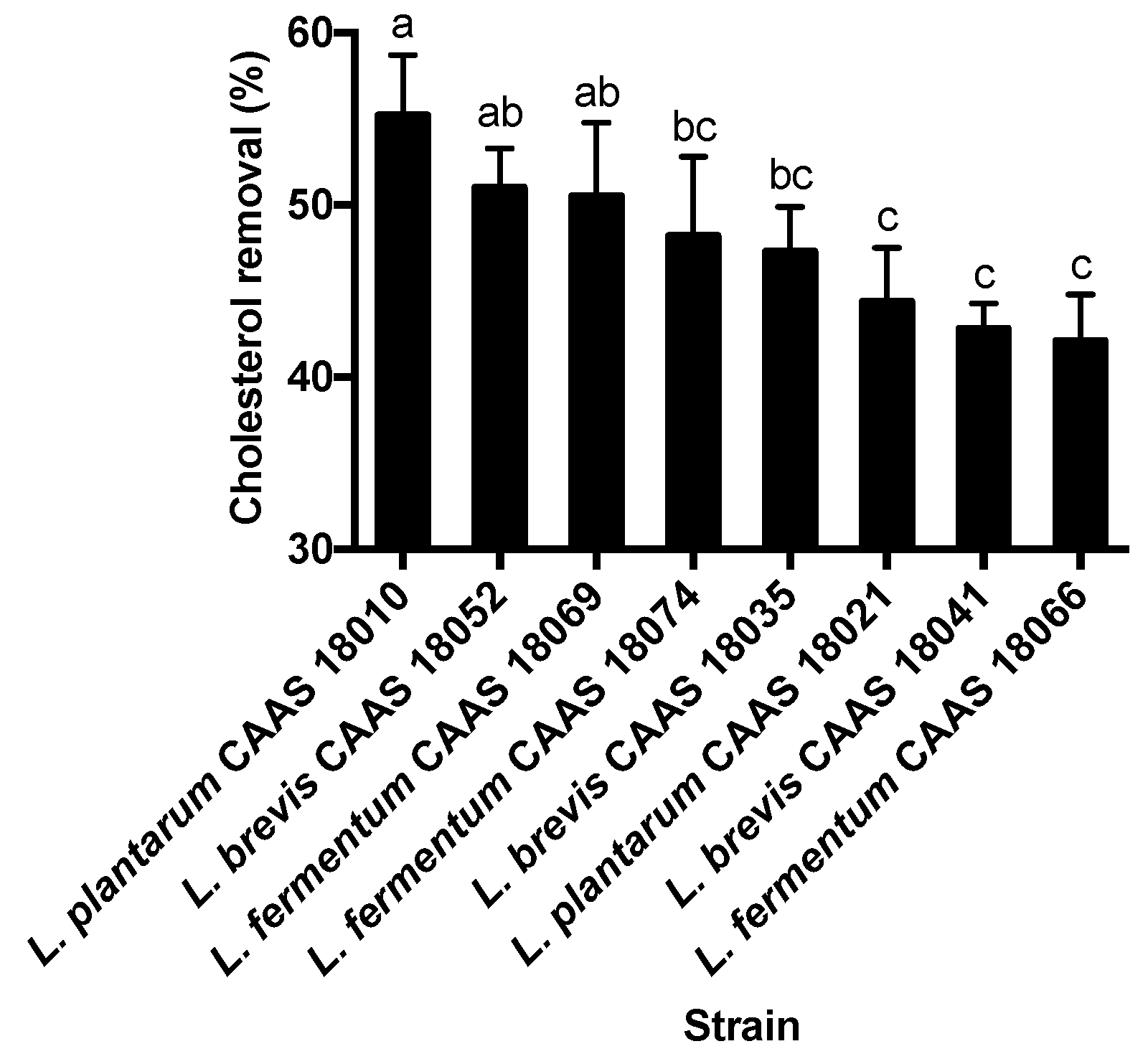
2.1. Cholesterol Removal
2.2. NPC1L1 Protein Down-Regulation
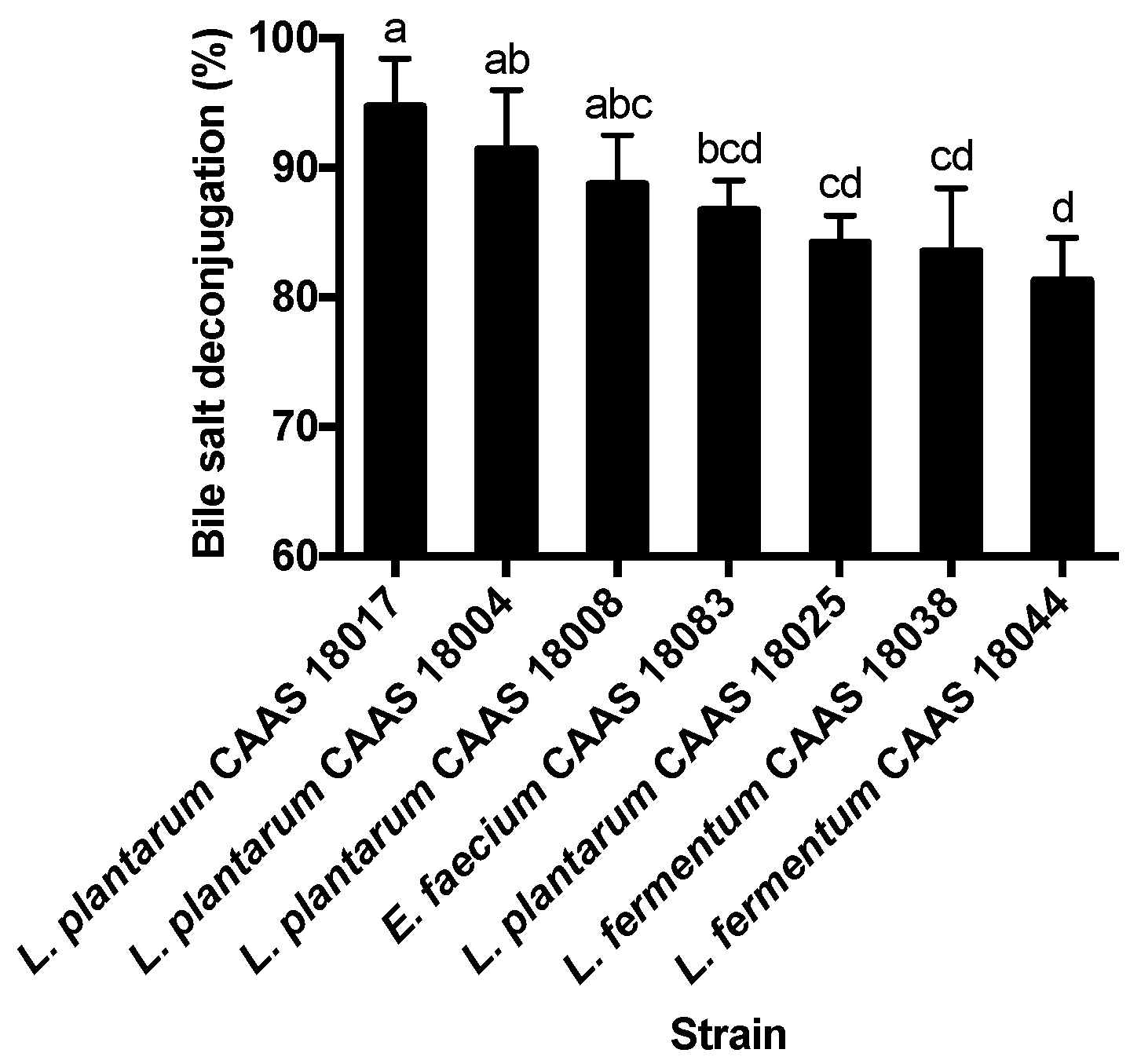
2.3. Bile Salt Deconjugation
2.4. Basic Probiotic Properties
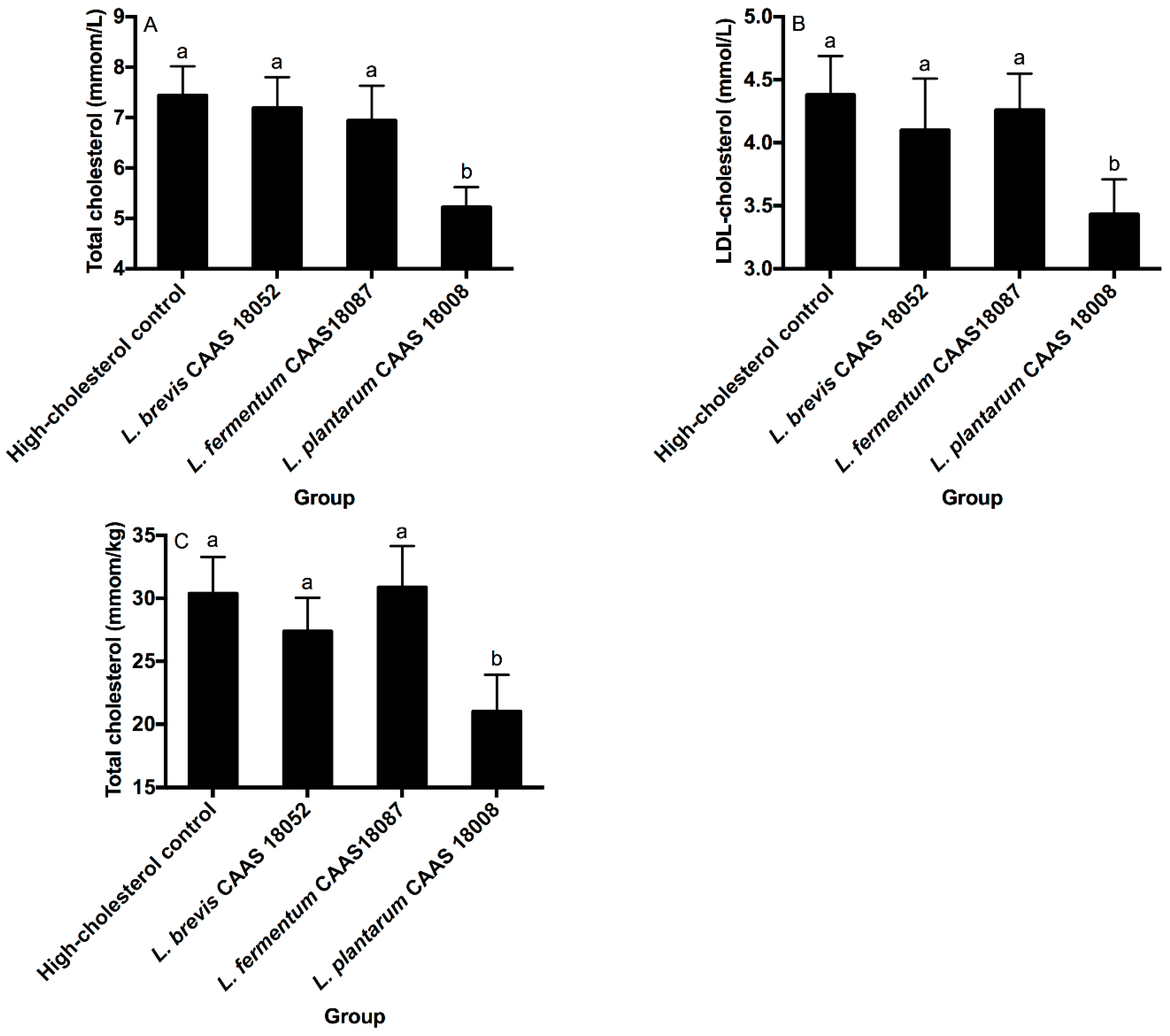
2.5. Serum and Hepatic Cholesterol
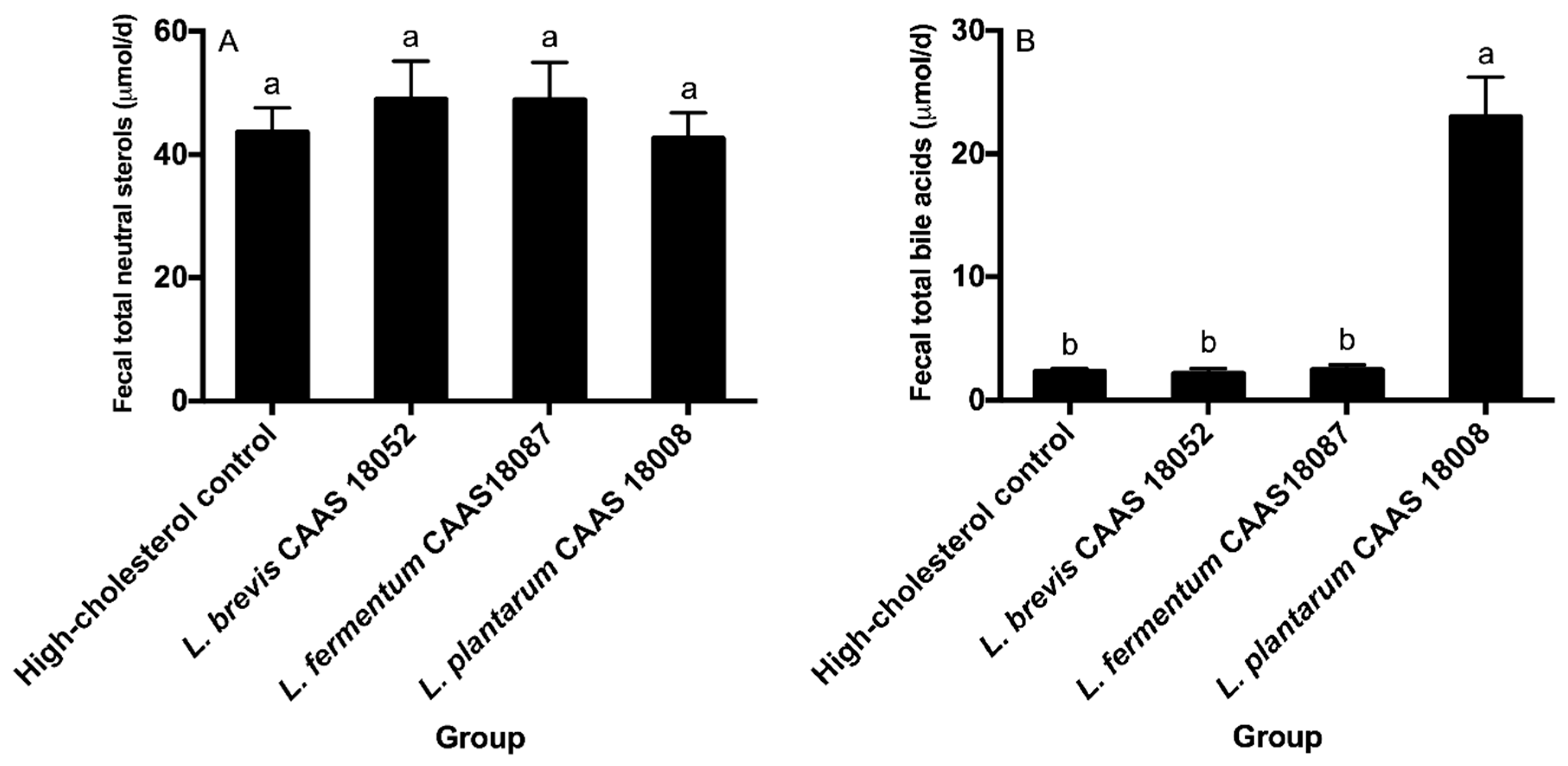
2.6. Fecal Sterol Excretion
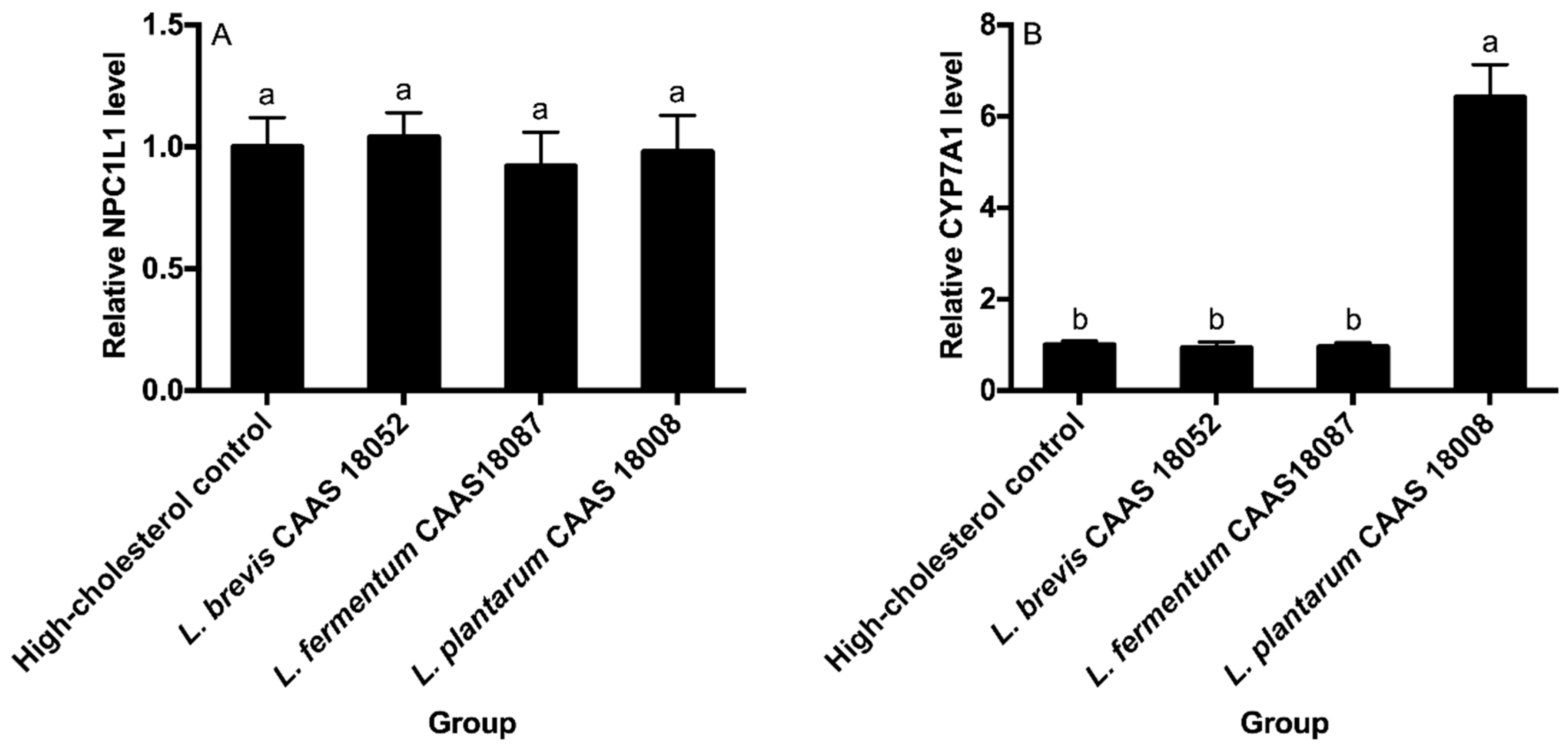
2.7. Small Intestinal NPC1L1 Protein and Hepatic Cholesterol-7α-Hydroxylase (CYP7A1)
3. Discussion
4. Materials and Methods
4.1. Source and Maintenance of Lactic Acid Bacteria Strains
4.2. Cholesterol Removal
4.3. NPC1L1 Proteiin Down-Regulation
4.4. Bile Salt Deconjugation
4.5. Determination of Acid Tolerance
4.6. Determination of Bile Tolerance
4.7. Determination of Adhesion Ability
4.8. Animal Feeding Trial
4.9. Determination of Serum and Hepatic Cholesterol Levels
4.10. Determination of Fecal Neutral and Acidic Sterols
4.11. Determination of Small Intestinal NPC1L1 Protein and Hepatic Cholesterol-7α-Hydroxylase (CYP7A1)
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- DiRienzo, D.B. Effect of probiotics on biomarkers of cardiovascular disease: implications for heart-healthy diets. Nutr. Rev. 2014, 72, 18–29. [Google Scholar] [CrossRef]
- Bartley, G.E.; Yokoyama, W.; Young, S.A.; Anderson, W.H.K.; Hung, S.-C.; Albers, D.R.; Langhorst, M.L.; Kim, H. Hypocholesterolemic effects of hydroxypropyl methylcellulose are mediated by altered gene expression in hepatic bile and cholesterol pathways of male hamsters. J. Nutr. 2010, 140, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Y.; Wu, S.-C.; Ng, C.-C.; Shyu, Y.-T. Effect of Lactobacillus-fermented adlay-based milk on lipid metabolism of hamsters fed cholesterol-enriched diet. Food Res. Int. 2010, 43, 819–824. [Google Scholar] [CrossRef]
- Howarth, G.S.; Wang, H. Role of endogenous microbiota, probiotics and their biological products in human health. Nutrients 2013, 5, 58–81. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-H.; Hsueh, Y.-H.; Kuo, J.-M.; Liu, S.-J. Characterization of a potential probiotic Lactobacillus brevis RK03 and efficient production of γ-aminobutyric acid in batch fermentation. Int. J. Mol. Sci. 2018, 19, 143. [Google Scholar] [CrossRef]
- Pavli, F.; Tassou, C.; Nychas GJ, E.; Chorianopoulos, N. Probiotic incorporation in edible films and coatings: bioactive solution for functional foods. Int. J. Mol. Sci. 2018, 19, 150. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, N.; Xi, A.; Ahmed, Z.; Zhang, B.; Bai, X. Effects of Lactobacillus plantarum MA2 isolated from Tibet kefir on lipid metabolism and intestinal microflora of rats fed on high-cholesterol diet. Appl. Microbiol. Biotechnol. 2009, 84, 341–347. [Google Scholar] [CrossRef]
- Ding, W.R.; Shi, C.; Chen, M.; Zhou, J.W.; Long, R.J.; Guo, X.S. Screening for lactic acid bacteria in traditional fermented Tibetan yak milk and evaluating their probiotic and cholesterol-lowering potentials in rats fed a high-cholesterol diet. J. Funct. Foods 2017, 32, 324–332. [Google Scholar] [CrossRef]
- Fuentes, M.C.; Lajo, T.; Carrioon, J.M.; Cune, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef]
- Jones, M.L.; Martoni, C.J.; Parent, M.; Prakash, S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Br. J. Nutr. 2012, 107, 1505–1513. [Google Scholar] [CrossRef]
- Lye, H.-S.; Rahmat-Ali, G.R.; Liong, M.-T. Mechanisms of cholesterol removal by lactobacilli under conditions that mimic the human gastrointestinal tract. Int. Dairy J. 2010, 20, 169–175. [Google Scholar] [CrossRef]
- Lye, F.S.; Alias, K.A.; Rusul, G.; Liong, M.T. Ultrasound treatment enhances cholesterol removal ability of lactobacilli. Ultrason. Sonochem. 2012, 19, 632–641. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, J.; Quan, G.; Wang, X.; Yang, L.; Zhong, L. Lactobacillus acidophilus ATCC 4356 prevents atherosclerosis via inhibition of intestinal cholesterol absorption in apolipoprotein E-knockout mice. Appl. Environ. Microbiol. 2014, 80, 7496–7504. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Tomaro-Duchesneau, C.; Martoni, C.J.; Prakash, S. Cholesterol lowering with bile salt hydrolase-active probiotic bacteria, mechanism of action, clinical evidence, and future direction for heart health applications. Expert Opin. Biol. Ther. 2013, 13, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.A.; Chang, H.C. Cholesterol-lowering effects of a putative probiotic strain Lactobacillus plantarum EM isolated from kimchi. Lwt-Food Sci. Technol. 2015, 62, 210–217. [Google Scholar] [CrossRef]
- Mandal, V.; Sen, S.K.; Mandal, N.C. Effect of prebiotics on bacteriocin production and cholesterol lowering activity of Pediococcus acidilactici LAB 5. World J. Microbiol. Biotechnol. 2009, 25, 1837–1847. [Google Scholar] [CrossRef]
- Kuda, T.; Yazaki, T.; Ono, M.; Takahashi, H.; Kimura, B. In vitro cholesterol-lowering properties of Lactobacillus plantarum AN6 isolated from aji-narezushi. Lett. Appl. Microbiol. 2013, 57, 187–192. [Google Scholar] [CrossRef]
- Guo, C.-F.; Zhao, D.; Yuan, Y.-H.; Yue, T.-L.; Liu, B.; Li, J.-Y. Lactobacillus casei-fermented milk improves serum and hepatic lipid profiles in diet-induced hypercholesterolaemic hamsters. J. Funct Foods 2016, 26, 691–697. [Google Scholar] [CrossRef]
- Ferraretto, L.F.; Shaver, R.D.; Luck, B.D. Silage review: Recent advances and future technologies for whole-plant and fractionated corn silage harvesting. J. Dairy Sci. 2018, 101, 3937–3951. [Google Scholar] [CrossRef]
- Pang, H.; Qin, G.; Tan, Z.; Li, Z.; Wang, Y.; Cai, Y. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and recA gene analysis. Syst. Appl. Microbiol. 2011, 34, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Ni, K.; Pang, H.; Wang, Y.; Cai, Y.; Jin, Q. Identification and antimicrobial activity detection of lactic acid bacteria isolated from corn stover silage. Asian Australas. J. Anim. Sci. 2015, 28, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhen, H.; Zhang, X.; Wang, S.; Zhang, Y.; Fang, Z.; Huang, Z.; Shi, P. Probiotic properties of Enterococcus strains isolated from the silage. J. Food Saf. 2015, 35, 108–118. [Google Scholar] [CrossRef]
- Weinberg, Z.G.; Chen, Y.; Gamburg, M. The passage of lactic acid bacteria from silage into rumen fluid, in vitro studies. J. Dairy Sci. 2004, 87, 3386–3397. [Google Scholar] [CrossRef]
- Dunne, C.; O’Mahony, L.; Murphy, L.; Thornton, G.; Morrissey, D.; O’Halloran, S.; Feeney, M.; Flynn, S.; Fitzgerald, G.; Daly, C.; et al. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 2001, 73, 386S–392S. [Google Scholar] [CrossRef]
- Noh, D.O.; Kim, S.H.; Gilliland, S.E. Incorporation of cholesterol into the cellular membrane of Lactobacillus acidophilus ATCC 43121. J. Dairy Sci. 1997, 80, 3107–3113. [Google Scholar] [CrossRef]
- Iranmanesh, M.; Ezzatpanah, H.; Mojgani, N. Antibacterial activity and cholesterol assimilation of lactic acid bacteria isolated from traditional Iranian dairy products. Lwt-Food Sci. Technol. 2014, 58, 355–359. [Google Scholar] [CrossRef]
- Lye, H.S.; Rusul, G.; Liong, M.T. Removal of cholesterol by lactobacilli via incorporation and conversion to coprostanol. J. Dairy Sci. 2010, 93, 1383–1392. [Google Scholar] [CrossRef]
- Liong, M.T.; Shah, N.P. Acid and bile tolerance and cholesterol removal ability of lactobacilli strains. J. Dairy Sci. 2005, 88, 55–66. [Google Scholar] [CrossRef]
- Kockx, M.; Kritharides, L. Intestinal cholesterol absorption. Curr. Opin. Lipidol. 2018, 29, 484–485. [Google Scholar] [CrossRef]
- McConnell, E.L.; Fadda, H.M.; Basit, A.W. Gut instincts: Explorations in intestinal physiology and drug delivery. Int. J. Pharm. 2008, 364, 213–226. [Google Scholar] [CrossRef]
- Altmann, S.W.; Davis, H.R.; Zhu, L.-J.; Yao, X.; Hoos, L.M.; Tetzloff, G.; Iyer, S.P.N.; Maguire, M.; Golovko, A.; Zeng, M.; et al. Niemann-Pick C1 like 1 protein is critical for intestinal cholesterol absorption. Science 2004, 303, 1201–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, B.-B.; Ge, L.; Li, B.-L.; Yan, Y.; Song, B.-L. Membrane topology of human NPC1L1, a key protein in enterohepatic cholesterol absorption. J. Lipid Res. 2009, 50, 1653–1662. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, J.F.; Cheng, Y.; Zheng, Y.C. The hypocholesterolaemic effects of Lactobacillus acidophilus American Type Culture Collection 4356 in rats are mediated by the down-regulation of Niemann-Pick C1-Like 1. Br. J. Nutr. 2010, 104, 807–812. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, Y. The probiotic Lactobacillus acidophilus reduces cholesterol absorption through the down-regulation of Niemann-Pick C1-like 1 in Caco-2 cells. Br. J. Nutr. 2010, 103, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-Y.; Ma, K.Y.; Liang, Y.; Peng, C.; Zuo, Y. Role and classification of cholesterol-lowering functional foods. J Funct Foods 2011, 3, 61–69. [Google Scholar] [CrossRef]
- Patel, A.K.; Singhania, R.R.; Pandey, A.; Chincholkar, S.B. Probiotic bile salt hydrolase: current developments and perspectives. Appl. Biochem. Biotechnol. 2010, 162, 166–180. [Google Scholar] [CrossRef]
- Sridevi, N.; Vishwe, P.; Prabhune, A. Hypocholesteremic effect of bile salt hydrolase from Lactobacillus buchneri ATCC 4005. Food Res. Int 2009, 42, 516–520. [Google Scholar] [CrossRef]
- Guo, C.-F.; Zhang, S.; Yuan, Y.-H.; Li, J.-Y.; Yue, T.-L. Bile salt hydrolase and S-layer protein are the key factors affecting the hypocholesterolemic activity of Lactobacillus casei-fermented milk in hamsters. Mol. Nutr. Food Res. 2018, 62, 1800728. [Google Scholar] [CrossRef]
- Gu, J.J.; Hofmann, A.F.; Tonnu, H.T.; Schteingart, C.D.; Mysels, K.J. Solubility of calcium salts of unconjugated and conjugated natural bile-acids. J. Lipid Res. 1992, 33, 635–646. [Google Scholar]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef]
- Ru, X.; Zhang, C.-C.; Yuan, Y.-H.; Yue, T.-L.; Guo, C.-F. Bile salt hydrolase activity is present in nonintestinal lactic acid bacteria at an intermediate level. Appl. Microbiol. Biotechnol. 2019, 103, 893–902. [Google Scholar] [CrossRef]
- Settanni, L.; Di Grigoli, A.; Tornambé, G.; Bellina, V.; Francesca, N.; Moschetti, G.; Bonanno, A. Persistence of wild Streptococcus thermophilus strains on wooden vat and during the manufacture of a traditional Caciocavallo type cheese. Int. J. Food Microbiol. 2012, 155, 73–81. [Google Scholar] [CrossRef]
- Fletouris, D.J.; Botsoglou, N.A.; Psomas, I.E.; Mantis, A.I. Rapid determination of cholesterol in milk and milk products by direct saponification and capillary gas chromatography. J Dairy Sci. 1998, 81, 2833–2840. [Google Scholar] [CrossRef]
- Tanaka, H.; Hashiba, H.; Kok, J.; Mierau, I. Bile salt hydrolase of Bifidobacterium longum - Biochemical and genetic characterization. Appl. Environ. Microbiol. 2000, 66, 2502–2512. [Google Scholar] [CrossRef]
- Hu, P.-L.; Yuan, Y.-H.; Yue, T.-L.; Guo, C.-F. A new method for the in vitro determination of the bile tolerance of potentially probiotic lactobacilli. Appl. Microbiol. Biotechnol. 2018, 102, 1903–1910. [Google Scholar] [CrossRef]
- Han, Q.; Kong, B.; Chen, Q.; Sun, F.; Zhang, H. In vitro comparison of probiotic properties of lactic acid bacteria isolated from Harbin dry sausages and selected probiotics. J. Funct. Foods 2017, 32, 391–400. [Google Scholar] [CrossRef]
- Pan, X.; Chen, F.; Wu, T.; Tang, H.; Zhao, Z. The acid, bile tolerance and antimicrobial property of Lactobacillus acidophilus NIT. Food Control 2009, 20, 598–602. [Google Scholar] [CrossRef]
- NIH (National Institutes of Health). Guide for the Care and Use of Laboratory Animals; National Academy Press: Washington, DC, USA, 1996. [Google Scholar]
- Guo, C.-F.; Yuan, Y.-H.; Yue, T.-L.; Li, J.-Y. Hamsters are a better model system than rats for evaluating the hypocholesterolemic efficacy of potential probiotic strains. Mol. Nutr. Food Res. 2018, 62, 1800170. [Google Scholar] [CrossRef]

| Hypothesized Pathway | Strain | Acid Tolerance (%) | Bile Tolerance (%) | Adhesion (Bacterial Counts /100 cells) |
|---|---|---|---|---|
| Greater cholesterol removal | L. plantarum CAAS 18010 | 61.6 ± 2.2 b | NG | 282 ± 30 a |
| L. plantarum CAAS 18021 | 33.6 ± 4.1 f | 83.6 ± 5.5 a | 281 ± 19 a | |
| L. brevis CAAS 18035 | 66.3 ± 2.0 ab | 41.5 ± 3.8 h | 251 ± 10 ab | |
| L. brevis CAAS 18041 | 65.9 ± 3.7 ab | 81.9 ± 5.6 ab | 228± 34 bc | |
| L. brevis CAAS 18052 | 65.2 ± 3.9 ab | 75.8 ± 2.8 bc | 224 ± 23 bc | |
| L. fermentum CAAS 18066 | 22.5 ± 1.0 h | 54.6 ± 4.1 g | 213± 28 bc | |
| L. fermentum CAAS 18069 | 55.5 ± 3.2 c | NG | 107 ± 19 e | |
| L. fermentum CAAS 18074 | 21.7 ± 3.0 h | 76.0 ± 5.1 bc | 241 ± 26 ab | |
| NPC1L1 protein down-regulation | L. fermentum CAAS 18062 | 30.8 ± 2.4 fg | 75.5 ± 4.2 bc | 245 ± 27 ab |
| L. fermentum CAAS 18067 | 49.9 ± 3.8 cd | 54.6 ± 3.6 g | 106 ± 19 e | |
| L. fermentum CAAS 18070 | 63.4± 2.9 ab | NG | 151 ± 23 d | |
| L. fermentum CAAS 18078 | 61.2 ± 3.4 b | 65.8 ± 2.3 de | 240 ± 22 abc | |
| Greater bile salt deconjugation | L. plantarum CAAS 18004 | 61.5 ± 3.4 b | NG | 205 ± 26 bc |
| L. plantarum CAAS 18008 | 68.5 ± 3.0 a | 75.7 ± 4.2 bc | 241± 14 ab | |
| L. plantarum CAAS 18017 | 55.5 ± 4.3 c | 80.7 ± 4.7 ab | 93 ± 20 e | |
| L. plantarum CAAS 18025 | 40.2 ± 3.0 e | 63.5 ± 4.2 ef | 101 ± 24 e | |
| L. brevis CAAS 18038 | 27.0 ± 2.6 gh | 71.8 ± 2.9 cd | 207± 29 bc | |
| L. brevis CAAS 18044 | 63.9 ± 3.9 ab | 74.8 ± 4.3 bc | 194 ± 34 c | |
| E. faecium CAAS 18083 | 45.5 ± 3.3 de | 57.5 ± 1.7 fg | 121 ± 26 de |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Zhang, S.; Lu, J.; Zhang, C.; Pang, X.; Lv, J. Screening for Cholesterol-Lowering Probiotics from Lactic Acid Bacteria Isolated from Corn Silage Based on Three Hypothesized Pathways. Int. J. Mol. Sci. 2019, 20, 2073. https://doi.org/10.3390/ijms20092073
Ma C, Zhang S, Lu J, Zhang C, Pang X, Lv J. Screening for Cholesterol-Lowering Probiotics from Lactic Acid Bacteria Isolated from Corn Silage Based on Three Hypothesized Pathways. International Journal of Molecular Sciences. 2019; 20(9):2073. https://doi.org/10.3390/ijms20092073
Chicago/Turabian StyleMa, Changlu, Shuwen Zhang, Jing Lu, Cai Zhang, Xiaoyang Pang, and Jiaping Lv. 2019. "Screening for Cholesterol-Lowering Probiotics from Lactic Acid Bacteria Isolated from Corn Silage Based on Three Hypothesized Pathways" International Journal of Molecular Sciences 20, no. 9: 2073. https://doi.org/10.3390/ijms20092073
APA StyleMa, C., Zhang, S., Lu, J., Zhang, C., Pang, X., & Lv, J. (2019). Screening for Cholesterol-Lowering Probiotics from Lactic Acid Bacteria Isolated from Corn Silage Based on Three Hypothesized Pathways. International Journal of Molecular Sciences, 20(9), 2073. https://doi.org/10.3390/ijms20092073







