Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato
Abstract
:1. Introduction
2. Results
2.1. Overview of Transcriptome Sequencing and Mapping to the Reference Genome
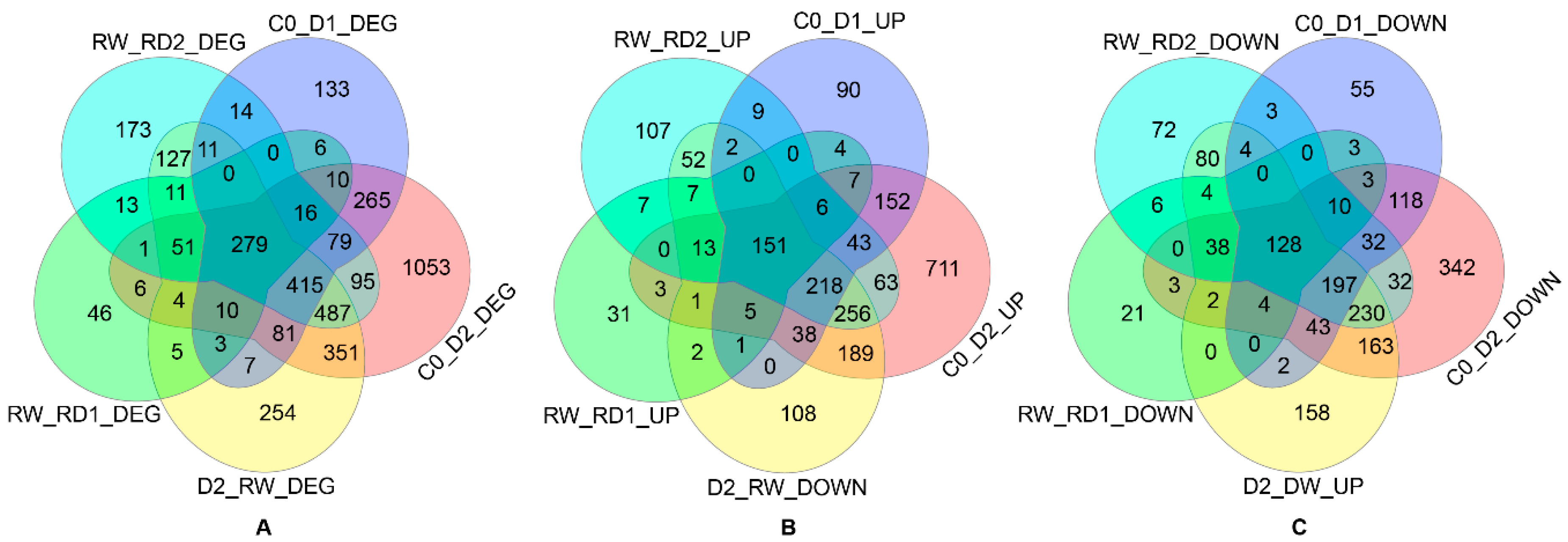
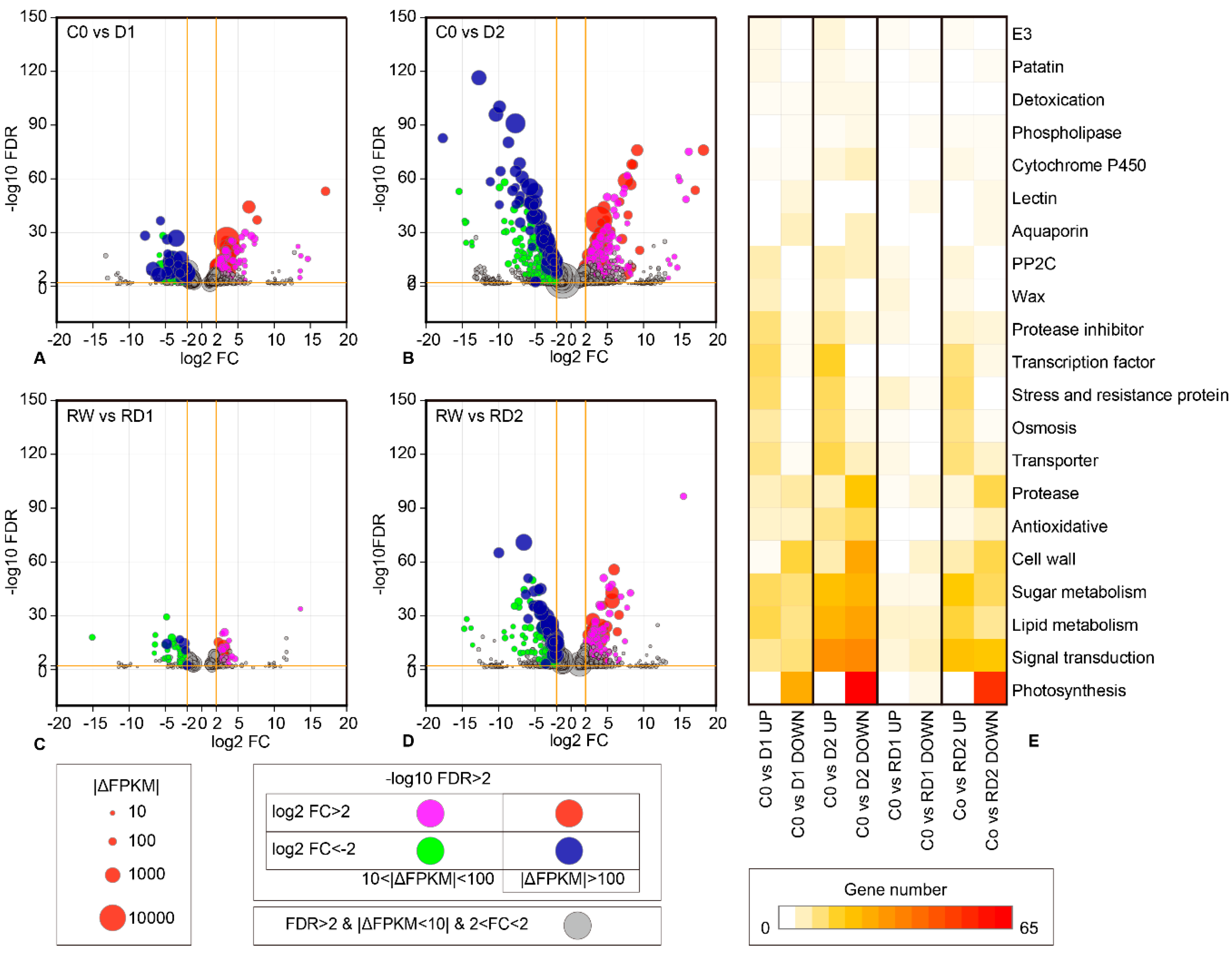
2.2. Differential Expression Analysis of Genes Responding to Dehydration, Rehydration and Re-Dehydration
2.3. GO Enrichment of DEG
2.4. KEGG Enrichment of DEGs
2.5. Drought-Responsive Genes
2.6. DEGs after Rehydration
2.7. Drought-Hardened DEGs
2.8. Real-Time qRT-PCR Verification of DEGs
3. Discussion
3.1. Perception and Transmission of Drought Stress Signals
3.2. Wax Synthesis and Transport Involved Drought Resistance in Potato
3.3. Photosynthesis Was Hardened by Drought in Potato
3.4. Drought Hardening
4. Materials and Methods
4.1. Plant Material
4.2. RNA-Seq
4.3. Alignment of RNA-seq Reads onto Reference Genome and Expression Analysis
4.4. GO and KEGG Enrichment
4.5. qRT-PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stokstad, E. The new potato. Breeders seek a breakthrough to help farmers facing an uncertain future. Science 2019, 363, 574–577. [Google Scholar] [CrossRef] [PubMed]
- Cabello, R.; Monneveux, P.; De Mendiburu, F.; Bonierbale, M. Comparison of yield based drought tolerance indices in improved varieties, genetic stocks and landraces of potato (Solanum tuberosum L.). Euphytica 2013, 193, 147–156. [Google Scholar] [CrossRef]
- Rykaczewska, K. Impact of heat and drought stresses on size and quality of the potato yield. Plant Soil Environ. 2017, 63, 40–46. [Google Scholar] [CrossRef] [Green Version]
- Monneveux, P.; Ramírez, D.A.; Pino, M.T. Drought tolerance in potato (S. tuberosum L.): Can we learn from drought tolerance research in cereals? Plant Sci. 2013, 205, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Muthoni, J.; Kabira, J.N. Potato production under drought conditions: Identification of adaptive traits. Int. J. Hortic. 2016, 6, 1–9. [Google Scholar] [CrossRef]
- Vasquez-Robinet, C.; Mane, S.P.; Ulanov, A.V.; Watkinson, J.I.; Stromberg, V.K.; De Koeyer, D.; Schafleitner, R.; Willmot, D.B.; Bonierbale, M.; Bohnert, H.J.; et al. Physiological and molecular adaptations to drought in Andean potato genotypes. J. Exp. Bot. 2008, 59, 2109–2123. [Google Scholar] [CrossRef] [Green Version]
- Evers, D.; Lefevre, I.; Legay, S.; Lamoureux, D.; Hausman, J.F.; Rosales, R.O.G.; Marca, L.R.T.; Hoffmann, L.; Bonierbale, M.; Schafleitner, R. Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J. Exp. Bot. 2010, 61, 2327–2343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Liu, B.; Ma, C.; Zhang, G.; Chang, J.; Si, H.; Wang, D. Transcriptome characterization and sequencing-based identification of drought-responsive genes in potato. Mol. Biol. Rep. 2014, 41, 505–517. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, H.; Gan, X.; Zhang, L.; Chen, Y.; Nie, F.; Shi, L.; Li, M.; Guo, Z.; Zhang, G.; et al. Transcriptome profiling of the potato (Solanum tuberosum L.) plant under drought stress and water-stimulus conditions. PLoS ONE 2015, 10, e0128041. [Google Scholar] [CrossRef]
- Yang, X.; Liu, J.; Xu, J.; Duan, S.; Wang, Q.; Li, G.; Jin, L. Transcriptome profiling reveals effects of drought stress on gene expression in diploid potato genotype P3–198. Int. J. Mol. Sci. 2019, 20, 852. [Google Scholar] [CrossRef] [Green Version]
- Pieczynski, M.; Wyrzykowska, A.; Milanowska, K.; Boguszewska-Mankowska, D.; Zagdanska, B.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Genomewide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnol. J. 2018, 16, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.L.; Chen, K.; Wang, S.S.; Gong, M. Osmoregulation as a key factor in drought hardening-induced drought tolerance in Jatropha curcas. Biol. Plant. 2015, 59, 529–536. [Google Scholar] [CrossRef]
- Li, P.; Yang, H.; Wang, L.; Liu, H.; Huo, H.; Zhang, C.; Liu, A.; Zhu, A.; Hu, J.; Lin, Y.; et al. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef] [PubMed]
- De Freitas Guedes, F.A.; Nobres, P.; Ferreira, D.C.R.; Menezes-Silva, P.E.; Ribeiro-Alves, M.; Correa, R.L.; DaMatta, F.M.; Alves-Ferreira, M. Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 2018, 147, 220–233. [Google Scholar] [CrossRef]
- Watkinson, J.I.; Hendricks, L.; Sioson, A.A.; Vasquez-Robinet, C.; Stromberg, V.; Heath, L.S.; Schuler, M.; Bohnert, H.J.; Bonierbale, M.; Grene, R. Accessions of Solanum tuberosum ssp. andigena show differences in photosynthetic recovery after drought stress as reflected in gene expression profiles. Plant Sci. 2006, 171, 745–758. [Google Scholar] [CrossRef]
- Ramírez, D.A.; Rolando, J.L.; Yactayo, W.; Monneveux, P.; Mares, V.; Quiroz, R. Improving potato drought tolerance through the induction of long-term water stress memory. Plant Sci. 2015, 238, 26–32. [Google Scholar] [CrossRef]
- Cabello, R.; De Mendiburu, F.; Bonierbale, M.; Monneveux, P.; Roca, W.; Chujoy, E. Large-scale evaluation of potato improved varieties, genetic stocks and landraces for drought tolerance. Am. J. Potato Res. 2012, 89, 400–410. [Google Scholar] [CrossRef]
- Huamán, Z.; Hawkes, J.G.; Rowe, P.R. Solanum ajanhuiri: An important diploid potato cultivated in the andean altiplano. Econ. Bot. 1980, 34, 335–343. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S. Arabidopsis duodecuple mutant of PYL ABA receptors reveals PYL repression of ABA-independent SnRK2 activity. Cell Rep. 2018, 23, 3340–3351. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, N.; Virlouvet, L.; Riethoven, J.J.; Fromm, M.; Avramova, Z. Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, T.; Zou, L.; Li, Y.; Yao, X.; Xu, F.; Deng, X.; Zhang, D.; Lin, H. Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 2018, 16, 2063–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klay, I.; Gouia, S.; Liu, M.; Mila, I.; Khoudi, H.; Bernadac, A.; Bouzayena, M.; Pirrello, J. Ethylene Response Factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018, 274, 137–145. [Google Scholar] [CrossRef]
- Hussain, S.S.; Kayani, M.A.; Amjad, M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol. Prog. 2011, 27, 297–306. [Google Scholar] [CrossRef]
- Singh, D.; Laxmi, A. Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Mane, S.P.; Robinet, C.V.; Ulanov, A.; Schafleitner, R.; Tincopa, L.; Gaudin, A.; Nomberto, G.; Alvarado, C.; Solis, C.; Bolivar, L.A. Molecular and physiological adaptation to prolonged drought stress in the leaves of two Andean potato genotypes. Funct. Plant Biol. 2008, 35, 669–688. [Google Scholar] [CrossRef]
- André, C.M.; Schafleitner, R.; Legay, S.; Lefèvre, I.; Aliaga, C.A.A.; Nomberto, G.; Hoffmann, L.; Hausman, J.F.; Larondelle, Y.; Evers, D. Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 2009, 70, 1107–1116. [Google Scholar] [CrossRef]
- Sprenger, H.; Kurowsky, C.; Horn, R.; Erban, A.; Seddig, S.; Rudack, K.; Fischer, A.; Zuther, H.; Köhl, K.; Hincha, D.K.; et al. The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ. 2016, 39, 2370–2389. [Google Scholar] [CrossRef]
- DeBono, A.; Yeats, T.H.; Rose, J.K.; Bird, D.; Jetter, R.; Kunst, L.; Samuels, L. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 2009, 21, 1230–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Lee, S.B.; Kim, H.J.; Min, M.K.; Hwang, I.; Suh, M.C. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1391–1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterk, P.; Booij, H.; Schellekens, G.A.; Van Kammen, A.; De Vries, S.C. Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell 1991, 3, 907–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerstiens, G. Cuticular water permeability and its physiological significance. J. Exp. Bot. 1996, 47, 1813–1832. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Guo, J.; Zhu, J.; Zhou, C. Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiol. Biochem. 2014, 75, 24–35. [Google Scholar] [CrossRef]
- Lee, J.; Yang, K.; Lee, M.; Kim, S.; Kim, J.; Lim, S.; Kang, G.H.; Min, S.R.; Kim, S.J.; Park, S.U.; et al. Differentiated cuticular wax content and expression patterns of cuticular wax biosynthetic genes in bloomed and bloomless broccoli (Brassica oleracea var. italica). Process Biochem. 2015, 50, 456–462. [Google Scholar] [CrossRef]
- Nott, A.; Jung, H.S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Kay, S.A. An expanding universe of circadian networks in higher plants. Trends Plant Sci. 2010, 15, 259–265. [Google Scholar] [CrossRef] [Green Version]
- Staneloni, R.J.; Rodriguez-Batiller, M.J.; Casal, J.J. Abscisic acid, high-light, and oxidative stress down-regulate a photosynthetic gene via a promoter motif not involved in phytochrome-mediated transcriptional regulation. Mol. Plant 2008, 1, 75–83. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Baker, E.A.; Procopiou, J. Effect of soil moisture status on leaf surface wax yield of some drought-resistant species. J. Hortic. Sci. 1980, 55, 85–87. [Google Scholar] [CrossRef]
- Banik, P.; Zeng, W.; Tai, H.; Bizimungu, B.; Tanino, K. Effects of drought acclimation on drought stress resistance in potato (Solanum tuberosum L.) genotypes. Environ. Exp. Bot. 2016, 126, 76–89. [Google Scholar] [CrossRef]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, B.; Li, C.; Lei, C.; Kong, C.; Yang, Y.; Gong, M. A comprehensive expression analysis of the expansin gene family in potato (Solanum tuberosum) discloses stress-responsive expansin-like B genes for drought and heat tolerances. PLoS ONE 2019, 14, e0219837. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The role of the late embryogenesis-abundant (LEA) protein family in development and the abiotic stress response: A comprehensive expression analysis of potato (Solanum tuberosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.; McQueen-Mason, S. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Lett. 2004, 559, 61–65. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, C.D.; Hamilton, J.P.; Childs, K.L.; Cepela, J.; Crisovan, E.; Vaillancourt, B.; Hirsch, C.N.; Habermann, M.; Neal, B.; Buell, C.R. Spud DB: A resource for mining sequences, genotypes, and phenotypes to accelerate potato breeding. Plant Genome 2014, 7. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]







| Comparison | DEG | Upregulation | Downregulation |
|---|---|---|---|
| C0–D1 | 1329 | 726 | 603 |
| C0–D2 | 3203 | 1857 | 1346 |
| D2–RW | 2096 | 1053 | 1043 |
| RW–RD1 | 461 | 239 | 222 |
| RW–RD2 | 1772 | 935 | 837 |
| C0–RW | 308 | 138 | 170 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato. Int. J. Mol. Sci. 2020, 21, 159. https://doi.org/10.3390/ijms21010159
Chen Y, Li C, Yi J, Yang Y, Lei C, Gong M. Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato. International Journal of Molecular Sciences. 2020; 21(1):159. https://doi.org/10.3390/ijms21010159
Chicago/Turabian StyleChen, Yongkun, Canhui Li, Jing Yi, Yu Yang, Chunxia Lei, and Ming Gong. 2020. "Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato" International Journal of Molecular Sciences 21, no. 1: 159. https://doi.org/10.3390/ijms21010159




