Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping
Abstract
:1. Introduction
2. Results
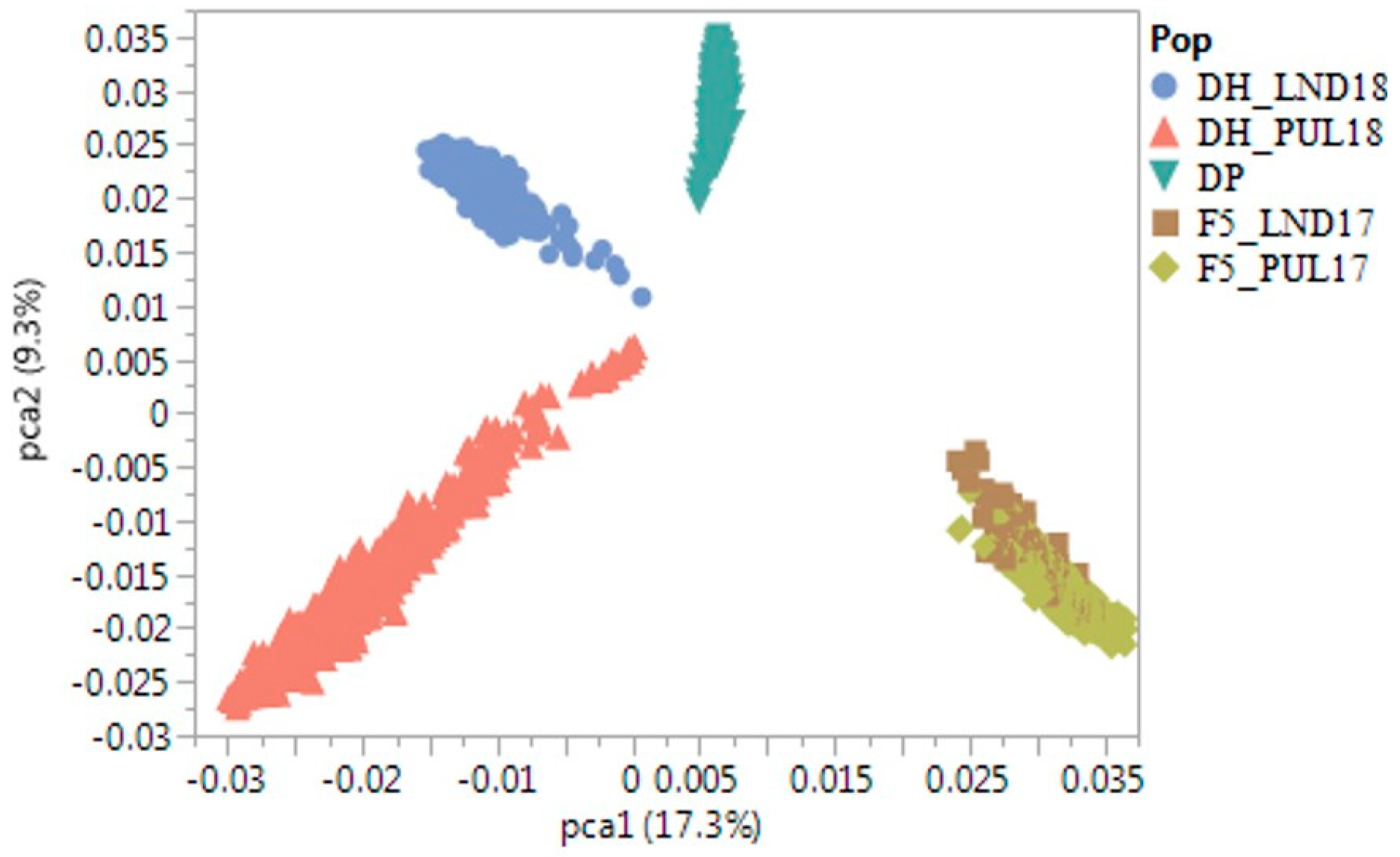
2.1. Phenotypic Correlations and Principal Components Analyses
2.2. Heritability, Genetic Correlations, and Response to Selection
2.3. Selection Based on Grain Yield and Spectral Reflectance Indices
2.4. Relationships between Actual and Predicted Grain Yield and Analysis of Principal Components
2.5. Prediction Accuracy for Grain Yield
3. Discussion
3.1. Indirect Selection for Grain Yield, Using Spectral Reflectance Indices
3.2. Heritability of the Spectral Reflectance Indices across Different Developmental Stages
3.3. Genetic and Phenotypic Correlations and Relative Selection Efficiency for Grain Yield
3.4. Genomic Prediction for Grain Yield
4. Materials and Methods
4.1. Plant Material
4.2. Collection and Analyses of Spectral Reflectance and Yield Data
4.3. Correlation between Spectral Reflectance Traits and Grain Yield
4.4. Predictive Models for Yield and Principal Components Analysis
4.5. SNP Marker Genotyping and Genomic Predictions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BLUE | Best linear unbiased estimate |
| BLUP | Best linear unbiased prediction |
| CV | Cross-validation |
| DH | Double haploid |
| DP | Diversity panel |
| HTP | High-throughput phenotyping |
| NDRE | Normalized difference red edge |
| NDVI | Normalized difference vegetative index |
| NWI-1 | Normalized water index 1 |
| NWI-2 | Normalized water index 2 |
| PCA | Principal components analysis |
| PNW | Pacific Northwest |
| QTL | Quantitative trait loci |
| RRBLUP | Ridge regression best linear unbiased prediction |
| SNP | Single nucleotide polymorphism |
| SR | Simple ratio |
| SRI | Spectral reflectance indices |
References
- Würschum, T. Modern field phenotyping opens new avenues for slection. In Applications of Genetic and Genomic Research in Cereals; Miedaner, T., Korzun, V., Eds.; Woodhead Publishing: Sawston, UK, 2018; pp. 164–171. [Google Scholar]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [Green Version]
- Montes, J.M.; Melchinger, A.E.; Reif, J.C. Novel throughput phenotyping platforms in plant genetic studies. Trends Plant Sci. 2007, 12, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Bosquet, L.; Crossa, J.; von Zitzewitz, J.; Serret, M.D.; Luis Araus, J. High-throughput phenotyping and genomic selection: The frontiers of crop breeding converge F. J. Integr. Plant Biol. 2012, 54, 312–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, A.J.; Berger, G.; Griffey, C.A.; Pitman, R.; Thomason, W.; Balota, M.; Ahmed, A. Genetic yield improvement in soft red winter wheat in the eastern United States from 1919 to 2009. Crop Sci. 2012, 52, 2097–2108. [Google Scholar] [CrossRef]
- Babar, M.A.; Reynolds, M.P.; van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Sci. 2006, 46, 1046–1057. [Google Scholar] [CrossRef]
- Gizaw, S.A.; Garland-Campbell, K.; Carter, A.H. Use of spectral reflectance for indirect selection of yield potential and stability in Pacific Northwest winter wheat. Field Crop Res. 2016, 196, 199–206. [Google Scholar] [CrossRef] [Green Version]
- Bowman, B.C.; Chen, J.; Zhang, J.; Wheeler, J.; Wang, Y.; Zhao, W.; Nayak, S.; Heslot, N.; Bockelman, H.; Bonman, J.M. Evaluating grain yield in spring wheat with canopy spectral reflectance. Crop Sci. 2015, 55, 1881–1890. [Google Scholar] [CrossRef] [Green Version]
- Kanke, Y.; Tubaña, B.; Dalen, M.; Harrell, D. Evaluation of red and red-edge reflectance-based vegetation indices for rice biomass and grain yield prediction models in paddy fields. Precis. Agric. 2016, 17, 507–530. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huang, W.; Peng, D.; Qin, Q.; Mortimer, H.; Casa, R.; Pignatti, S.; Laneve, G.; Pascucci, S.; et al. Vegetation indices combining the red and red-edge spectral information for leaf area index retrieval. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2018, 11, 1482–1493. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, N.; Villegas, D.; Casadesus, J.; Araus, J.L.; Royo, C. Spectral vegetation indices as nondestructive tools for determining durum wheat yield. Agron. J. 2000, 92, 83–91. [Google Scholar] [CrossRef]
- Babar, M.A.; Van Ginkel, M.; Klatt, A.R.; Prasad, B.; Reynolds, M.P. The potential of using spectral reflectance indices to estimate yield in wheat grown under reduced irrigation. Euphytica 2006, 150, 155–172. [Google Scholar] [CrossRef]
- Prasad, B.; Carver, B.F.; Stone, M.L.; Babar, M.A.; Raun, W.R.; Klatt, A.R. Genetic analysis of indirect selection for winter wheat grain yield using spectral reflectance indices. Crop Sci. 2007, 47, 1416–1425. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, M.; Reynolds, M.P.; Raun, W.R.; Stone, M.L.; Klatt, A.R. Spectral water indices for assessing yield in elite bread wheat genotypes under well-irrigated, water-Stressed, and high-temperature conditions. Crop Sci. 2010, 50, 197–214. [Google Scholar] [CrossRef]
- Thomson, M.J. High-throughput SNP genotyping to accelerate crop improvement. Plant Breed. Biotechnol. 2014, 2, 195–212. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Varshney, R.K.; Terauchi, R.; McCouch, S.R. Harvesting the promising fruits of genomics: Applying genome sequencing technologies to crop breeding. PLoS Biol. 2014, 12, e1001883. [Google Scholar] [CrossRef] [Green Version]
- Rasheed, A.; Xia, X. From markers to genome-based breeding in wheat. Theor. Appl. Genet. 2019, 132, 767–784. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueño, J.; Camacho-González, J.M.; Pérez-Elizalde, S.; Beyene, Y. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Mirdita, V.; He, S.; Zhao, Y.; Korzun, V.; Bothe, R.; Ebmeyer, E.; Reif, J.C.; Jiang, Y. Potential and limits of whole genome prediction of resistance to Fusarium head blight and Septoria tritici blotch in a vast Central European elite winter wheat population. Theor. Appl. Genet. 2015, 128, 2471–2481. [Google Scholar] [CrossRef]
- Arruda, M.P.; Brown, P.; Brown-Guedira, G.; Krill, A.M.; Thurber, C.; Merrill, K.R.; Foresman, B.J.; Kolb, F.L. Genome-wide association mapping of fusarium head blight resistance in wheat using genotyping-by-sequencing. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Würschum, T.; Leiser, W.L.; Langer, S.M.; Tucker, M.R.; Longin, C.F.H. Phenotypic and genetic analysis of spike and kernel characteristics in wheat reveals long-term genetic trends of grain yield components. Theor. Appl. Genet. 2018, 131, 2071–2084. [Google Scholar] [CrossRef] [PubMed]
- Rutkoski, J.; Poland, J.; Mondal, S.; Autrique, E.; Pérez, L.G.; Crossa, J.; Reynolds, M.; Singh, R. Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3 (Bethesda) 2016, 6, 2799–2808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Rutkoski, J.E.; Poland, J.A.; Crossa, J.; Jannink, J.-L.; Sorrells, M.E. Multitrait, random regression, or simple repeatability model in high-throughput phenotyping data improve genomic prediction for wheat grain yield. Plant Genome 2017, 10. [Google Scholar] [CrossRef] [Green Version]
- Crain, J.; Mondal, S.; Rutkoski, J.; Singh, R.P.; Poland, J. Combining high-throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. Plant Genome 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Juliana, P.; Singh, R.P.; Poland, J.; Mondal, S.; Crossa, J.; Montesinos-López, O.A.; Dreisigacker, S.; Pérez-Rodríguez, P.; Huerta-Espino, J.; Crespo-Herrera, L.; et al. Prospects and challenges of applied genomic selection—A new paradigm in breeding for grain yield in bread wheat. Plant Genome 2018, 11. [Google Scholar] [CrossRef] [Green Version]
- Ferrio, J.P.; Villegas, D.; Zarco, J.; Aparicio, N.; Araus, J.L.; Royo, C. Assessment of durum wheat yield using visible and near-infrared reflectance spectra of canopies. Field Crop. Res. 2005, 94, 126–148. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics; Oliver and Boyd: Edinburgh, UK; London, UK, 1960. [Google Scholar]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A rapid monitoring of NDVI across the wheat growth cycle for grain yield prediction using a multi-spectral UAV platform. Plant Sci. 2018, 282, 95–103. [Google Scholar] [CrossRef]
- Babar, M.A.; Reynolds, M.P.; Van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral reflectance indices as a potential indirect selection criteria for wheat yield under irrigation. Crop Sci. 2006, 46, 578–588. [Google Scholar] [CrossRef]
- Ma, B.L.; Dwyer, L.M.; Costa, C.; Cober, E.R.; Morrison, M.J. Early prediction of soybean yield from canopy reflectance measurements. Agron. J. 2001, 93, 1227–1234. [Google Scholar] [CrossRef] [Green Version]
- Prey, L.; von Bloh, M.; Schmidhalter, U. Evaluating RGB imaging and multispectral active and hyperspectral passive sensing for assessing early plant vigor in winter wheat. Sensors 2018, 18, 2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, R.E.; Addison, C.K.; Babar, A.; Acuna, A.; Lozada, D.; Subramanian, N.; Arguello, M.N.; Miller, R.G.; Brown-Guedira, G.; Guedira, M.; et al. Diagnostic markers for vernalization and photoperiod loci improve genomic selection for grain yield and spectral reflectance in wheat. Crop Sci. 2018, 58, 242–252. [Google Scholar] [CrossRef]
- Rutkoski, J.; Benson, J.; Jia, Y.; Brown-Guedira, G.; Jannink, J.-L.; Sorrells, M. Evaluation of Genomic Prediction Methods for Fusarium Head Blight Resistance in Wheat. Plant Genome 2012, 5, 51–61. [Google Scholar] [CrossRef] [Green Version]
- Bernardo, R. Genomewide Selection when Major Genes Are Known. Crop Sci. 2014, 54, 68–75. [Google Scholar] [CrossRef]
- Michel, S.; Kummer, C.; Gallee, M.; Hellinger, J.; Ametz, C.; Akgöl, B.; Epure, D.; Löschenberger, F.; Buerstmayr, H. Improving the baking quality of bread wheat by genomic selection in early generations. Theor. Appl. Genet. 2018, 131, 477–493. [Google Scholar] [CrossRef] [PubMed]
- Galiano-Carneiro, A.L.; Boeven, P.H.G.; Maurer, H.P.; Würschum, T.; Miedaner, T. Genome-wide association study for an efficient selection of Fusarium head blight resistance in winter triticale. Euphytica 2018, 215, 4. [Google Scholar] [CrossRef]
- Sarinelli, J.M.; Murphy, J.P.; Tyagi, P.; Holland, J.B.; Johnson, J.W.; Mergoum, M.; Mason, R.E.; Babar, A.; Harrison, S.; Sutton, R.; et al. Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor. Appl. Genet. 2019, 132, 1247. [Google Scholar] [CrossRef] [Green Version]
- Charmet, G.; Storlie, E.; Oury, F.X.; Laurent, V.; Beghin, D.; Chevarin, L.; Lapierre, A.; Perretant, M.R.; Rolland, B.; Heumez, E.; et al. Genome-wide prediction of three important traits in bread wheat. Mol. Breed. 2014, 34, 1843–1852. [Google Scholar] [CrossRef] [Green Version]
- Jarquin, D.; Specht, J.; Lorenz, A. Prospects of genomic prediction in the USDA soybean germplasm collection: Historical data creates robust models for enhancing selection of accessions. G3 Genes Genomes Genet. 2016, 6, 2329–2341. [Google Scholar] [CrossRef] [Green Version]
- Michel, S.; Ametz, C.; Gungor, H.; Epure, D.; Grausgruber, H.; Löschenberger, F.; Buerstmayr, H. Genomic selection across multiple breeding cycles in applied bread wheat breeding. Theor. Appl. Genet. 2016, 129, 1179–1189. [Google Scholar] [CrossRef] [Green Version]
- Poudel, H.P.; Sanciangco, M.D.; Kaeppler, S.M.; Buell, C.R.; Casler, M.D. Genomic prediction for winter survival of lowland switchgrass in the northern USA. G3 Genes Genomes Genet. 2019, 9, 1921–1931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battenfield, S.D.; Guzmán, C.; Gaynor, R.C.; Singh, R.P.; Peña, R.J.; Dreisigacker, S.; Fritz, A.K.; Poland, J.A. Genomic selection for processing and end-use quality traits in the CIMMYT spring bread wheat breeding program. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Lozada, D.N.; Mason, R.E.; Sarinelli, J.M.; Guedira, G.-B. Accuracy of genomic selection for grain yield and agronomic traits in soft red winter wheat. BMC Genet. 2019, 20, 82. [Google Scholar] [CrossRef] [PubMed]
- Lozada, D.N.; Carter, A.H. Accuracy of single and multi-trait genomic prediction models for grain yield in US Pacific Northwest winter wheat. Crop Breed. Genet. Genom. 2019, 1, e190012. [Google Scholar]
- Asoro, F.G.; Newell, M.A.; Beavis, W.D.; Scott, M.P.; Jannink, J.-L. Accuracy and training population design for genomic selection on quantitative traits in elite North American oats. Plant Genome 2011, 4, 132–144. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, A.J.; Smith, K.P. Adding genetically distant individuals to training populations reduces genomic prediction accuracy in barley. Crop Sci. 2015, 55, 2657–2667. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Wang, H.; Beyene, Y.; Semagn, K.; Liu, Y.; Cao, S.; Cui, Z.; Ruan, Y.; Burgueño, J.; San Vicente, F.; et al. Effect of trait heritability, training population size and marker density on genomic prediction accuracy estimation in 22 bi-parental tropical maize populations. Front. Plant. Sci. 2017, 8, 1916. [Google Scholar] [CrossRef]
- Sun, J.; Poland, J.A.; Mondal, S.; Crossa, J.; Juliana, P.; Singh, R.P.; Rutkoski, J.E.; Jannink, J.-L.; Crespo-Herrera, L.; Velu, G.; et al. High-throughput phenotyping platforms enhance genomic selection for wheat grain yield across populations and cycles in early stage. Theor. Appl. Genet. 2019, 132, 1705–1720. [Google Scholar] [CrossRef]
- Moore, J.K.; Manmathan, H.K.; Anderson, V.A.; Poland, J.A.; Morris, C.F.; Haley, S.D. Improving genomic prediction for pre-harvest sprouting tolerance in wheat by weighting large-effect quantitative trait loci. Crop Sci. 2017, 57, 1315–1324. [Google Scholar] [CrossRef]
- Huang, M.; Ward, B.; Griffey, C.; Van Sanford, D.; McKendry, A.; Brown-Guedira, G.; Tyagi, P.; Sneller, C. The accuracy of genomic prediction between environments and populations for soft wheat traits. Crop Sci. 2018, 58, 2274–2280. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Kurle, J.E.; Anderson, G.; Young, N.D. Association mapping and genomic prediction for resistance to sudden death syndrome in early maturing soybean germplasm. Mol. Breed. 2015, 35, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, S.B.; Dias, K.O.G.; Ferreira, D.F.; Brown, P.J. Efficiency of multi-trait, indirect, and trait-assisted genomic selection for improvement of biomass sorghum. Theor. Appl. Genet. 2018, 131, 747–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulthess, A.W.; Wang, Y.; Miedaner, T.; Wilde, P.; Reif, J.C.; Zhao, Y. Multiple-trait- and selection indices-genomic predictions for grain yield and protein content in rye for feeding purposes. Theor. Appl. Genet. 2016, 129, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Federer, W.T.; Raghavarao, D. On augmented designs. Biometrics 1975, 31, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Peterson, C.J.; Allan, R.E.; Rubenthaler, G.L.; Line, R.F. Registration of ‘Eltan’ wheat. Crop Sci. 1991, 31, 1704. [Google Scholar] [CrossRef]
- Allan, R.E.; Peterson, C.J.; Rubenthaler, G.L.; Line, R.F.; Roberts, D.E. Registration of ‘Madsen’ wheat. Crop Sci. 1989, 29, 1575–1576. [Google Scholar] [CrossRef]
- Jones, S.S.; Lyon, S.R.; Balow, K.A.; Gollnick, M.A.; Murphy, K.M.; Kuehner, J.S.; Murray, T.D.; Chen, X.M.; Engle, D.A.; Campbell, K.G. Registration of ’Xerpha’ wheat. J. Plant Regist. 2010, 4, 137–140. [Google Scholar] [CrossRef]
- Jones, S.S.; Murray, T.D.; Lyon, S.R.; Morris, C.F.; Line, R.F. Registration of ‘Bruehl’ wheat. (Registrations of Cultivars). Crop Sci. 2001, 41, 2006–2008. [Google Scholar] [CrossRef]
- Carter, A.H.; Jones, S.S.; Lyon, S.R.; Balow, K.A.; Shelton, G.B.; Higginbotham, R.W.; Chen, X.M.; Engle, D.A.; Baik, B.; Guy, S.O. Registration of ‘Otto’ wheat. J. Plant Regist. 2013, 7, 195–200. [Google Scholar] [CrossRef] [Green Version]
- Carter, A.H.; Jones, S.S.; Balow, K.A.; Shelton, G.B.; Burke, A.B.; Lyon, S.; Higginbotham, R.W.; Chen, X.M.; Engle, D.A.; Murray, T.D. Registration of ‘Jasper’ soft white winter wheat. J. Plant Regist. 2017, 11, 263–268. [Google Scholar] [CrossRef]
- Zemetra, R.S.; Souza, E.J.; Lauver, M.; Windes, J.; Guy, S.O.; Brown, B.; Robertson, L.; Kruk, M. Registration of ‘Brundage’ wheat. Crop Sci 1998, 38, 67. [Google Scholar] [CrossRef]
- Carter, A.H.; Jones, S.S.; Cai, X.; Lyon, S.R.; Balow, K.A.; Shelton, G.B.; Higginbotham, R.W.; Chen, X.M.; Engle, D.A.; Baik, B. Registration of ‘Puma’soft white winter wheat. J. Plant Regist. 2014, 8, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Zhao, W.; Zhou, D.; Gong, H. Sensitivity analysis of vegetation reflectance to biochemical and biophysical variables at leaf, canopy, and regional scales. IEEE Trans. Geosci. Remote Sens. 2014, 52, 4014–4024. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; Texas A&M University Remote Sensing Center: College Town, TX, USA, 1973. [Google Scholar]
- Stenberg, P.; Rautiainen, M.; Manninen, T.; Voipio, P.; Smolander, H. Reduced simple ratio better than NDVI for estimating LAI in finnish pine and spruce stands. Silva Fenn. 2004, 38, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, F.; Alvarado, G.; Pacheco, Á.; Burgueño, J. ACBD-R. Augmented Complete Block Design with R for Windows; Version 4.0; CIMMYT Research Data & Software Repository Network: Texcoco de Mora, Mexico, 2018. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- SAS Institute. SAS® 9.4 System Options Reference, 5th ed.; SAS Institute Inc.: Cary, NC, USA, 2016. [Google Scholar]
- Falconer, D.S. Introduction to Quantitative Genetics, 3rd ed.; Longman Scientific and Technical: New York, NY, USA, 1989. [Google Scholar]
- Poland, J.A.; Brown, P.J.; Sorrells, M.E.; Jannink, J.-L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 2012, 7, e32253. [Google Scholar] [CrossRef] [Green Version]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 1–13. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Money, D.; Gardner, K.; Migicovsky, Z.; Schwaninger, H.; Zhong, G.-Y.; Myles, S. LinkImpute: Fast and accurate genotype imputation for nonmodel organisms. G3 Genes Genomes Genet. 2015, 5, 2383–2390. [Google Scholar] [CrossRef] [Green Version]
- Roger, J.S. Measure of genetic similarity and genetic distance. In Studies in Genetics VII; University of Texas Publishing: Austin, TX, USA, 1972; pp. 145–153. [Google Scholar]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 250–255. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Zhang, Z. iPat: Intelligent prediction and association tool for genomic research. Bioinformatics 2018, 34, 1925–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heffner, E.; Jannink, J.-L.; Iwata, H.; Souza, E.; Sorrells, M.E. Genomic selection accuracy for grain quality traits in biparental wheat populations. Crop Sci. 2011, 51, 2597–2606. [Google Scholar] [CrossRef] [Green Version]



| Index 1 | Growth Stage 2 | Heritability | Mean | Correlation with Yield 3 | Response to Selection 4 | Correlated Response 5 | Relative Selection Efficiency 6 | |
|---|---|---|---|---|---|---|---|---|
| Phenotypic | Genetic | |||||||
| NDRE-1 | Hd | 0.26 | 0.75 | 0.77 | 0.64 | 0.05 | 0.025 | 0.74 |
| Gf1 | 0.63 | 0.71 | 0.77 | 0.65 | 0.37 | 0.039 | 1.17 | |
| Gf2 | 0.30 | 0.51 | 0.86 | 0.59 | 0.08 | 0.024 | 0.73 | |
| NDRE-2 | Hd | 0.42 | 0.14 | 0.78 | 0.65 | 0.10 | 0.032 | 0.96 |
| Gf1 | 0.53 | 0.13 | 0.73 | 0.65 | 0.12 | 0.036 | 1.07 | |
| Gf2 | 0.30 | 0.10 | 0.81 | 0.58 | 0.08 | 0.024 | 0.72 | |
| NDVI | Hd | 0.24 | 0.83 | 0.75 | 0.64 | 0.04 | 0.024 | 0.71 |
| Gf1 | 0.52 | 0.80 | 0.76 | 0.66 | 0.07 | 0.036 | 1.08 | |
| Gf2 | 0.37 | 0.63 | 0.86 | 0.58 | 0.08 | 0.027 | 0.80 | |
| NWI-1 | Hd | 0.23 | −0.08 | −0.63 | −0.51 | 0.04 | −0.018 | −0.56 |
| Gf1 | 0.16 | −0.07 | −0.61 | −0.42 | 0.02 | −0.013 | −0.38 | |
| Gf2 | 0.26 | −0.06 | −0.88 | −0.58 | 0.06 | −0.022 | −0.67 | |
| SR | Hd | 0.41 | 18.61 | 0.78 | 0.67 | 0.11 | 0.032 | 0.97 |
| Gf1 | 0.55 | 15.81 | 0.66 | 0.57 | 0.43 | 0.031 | 0.96 | |
| Gf2 | 0.31 | 7.24 | 0.82 | 0.57 | 0.07 | 0.024 | 0.72 | |
| Test Population | Diversity Panel | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| LND15 | LND17 | LND18 | LND_Com | PUL15 | PUL16 | PUL17 | PUL18 | PUL_Com | |
| F5_LND17 | −0.08 | 0.26 * | −0.06 | −0.03 | 0.11 | 0.19 | 0.24 | 0.22 | 0.19 |
| DH_LND18 | 0.07 | −0.03 | −0.08 | 0.004 | 0.003 | 0.02 | −0.02 | −0.04 | 0.02 |
| F5_PUL17 | −0.06 | −0.11 | −0.01 | −0.062 | −0.03 | 0.02 | −0.07 | −0.02 | −0.03 |
| DH_PUL18 | 0.01 | −0.01 | 0.04 | 0.03 | 0.04 | 0.09 | 0.06 | 0.07 | 0.05 |
| Index 1 | LND17 | LND18 | PUL17 | PUL18 |
|---|---|---|---|---|
| NDRE-1 | 66.1 | 47.0 | 29.6 | 29.6 |
| NDRE-2 | 66.1 | 46.1 | 26.1 | 31.3 |
| NDVI | 65.2 | 47.8 | 31.3 | 29.6 |
| NWI-1 | 66.1 | 50.4 | 13.9 | 30.4 |
| SR | 65.2 | 45.2 | 31.3 | 27.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozada, D.N.; Godoy, J.V.; Ward, B.P.; Carter, A.H. Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping. Int. J. Mol. Sci. 2020, 21, 165. https://doi.org/10.3390/ijms21010165
Lozada DN, Godoy JV, Ward BP, Carter AH. Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping. International Journal of Molecular Sciences. 2020; 21(1):165. https://doi.org/10.3390/ijms21010165
Chicago/Turabian StyleLozada, Dennis N., Jayfred V. Godoy, Brian P. Ward, and Arron H. Carter. 2020. "Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping" International Journal of Molecular Sciences 21, no. 1: 165. https://doi.org/10.3390/ijms21010165
APA StyleLozada, D. N., Godoy, J. V., Ward, B. P., & Carter, A. H. (2020). Genomic Prediction and Indirect Selection for Grain Yield in US Pacific Northwest Winter Wheat Using Spectral Reflectance Indices from High-Throughput Phenotyping. International Journal of Molecular Sciences, 21(1), 165. https://doi.org/10.3390/ijms21010165





