Insight into the Molecular Interaction of Cloxyquin (5-chloro-8-hydroxyquinoline) with Bovine Serum Albumin: Biophysical Analysis and Computational Simulation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Cloxyquin-Induced Fluorescence Quenching of BSA
2.2. BSA and Cloxyquin Absorption Spectra
2.3. Binding Parameters of BSA–Cloxyquin Complex
2.4. Fluorescence Resonance Energy Transfer (FRET) Investigation
2.5. Synchronous Fluorescence Analysis
2.6. Investigation of Conformational Changes
2.6.1. Three-Dimensional (3D) Spectrum Investigation and Dynamic Light Scattering (DLS) Analysis
2.6.2. Secondary Structure Examination by Circular Dichroism Spectroscopy
2.7. Probing the Binding Sites with Site Marker Displacement Studies
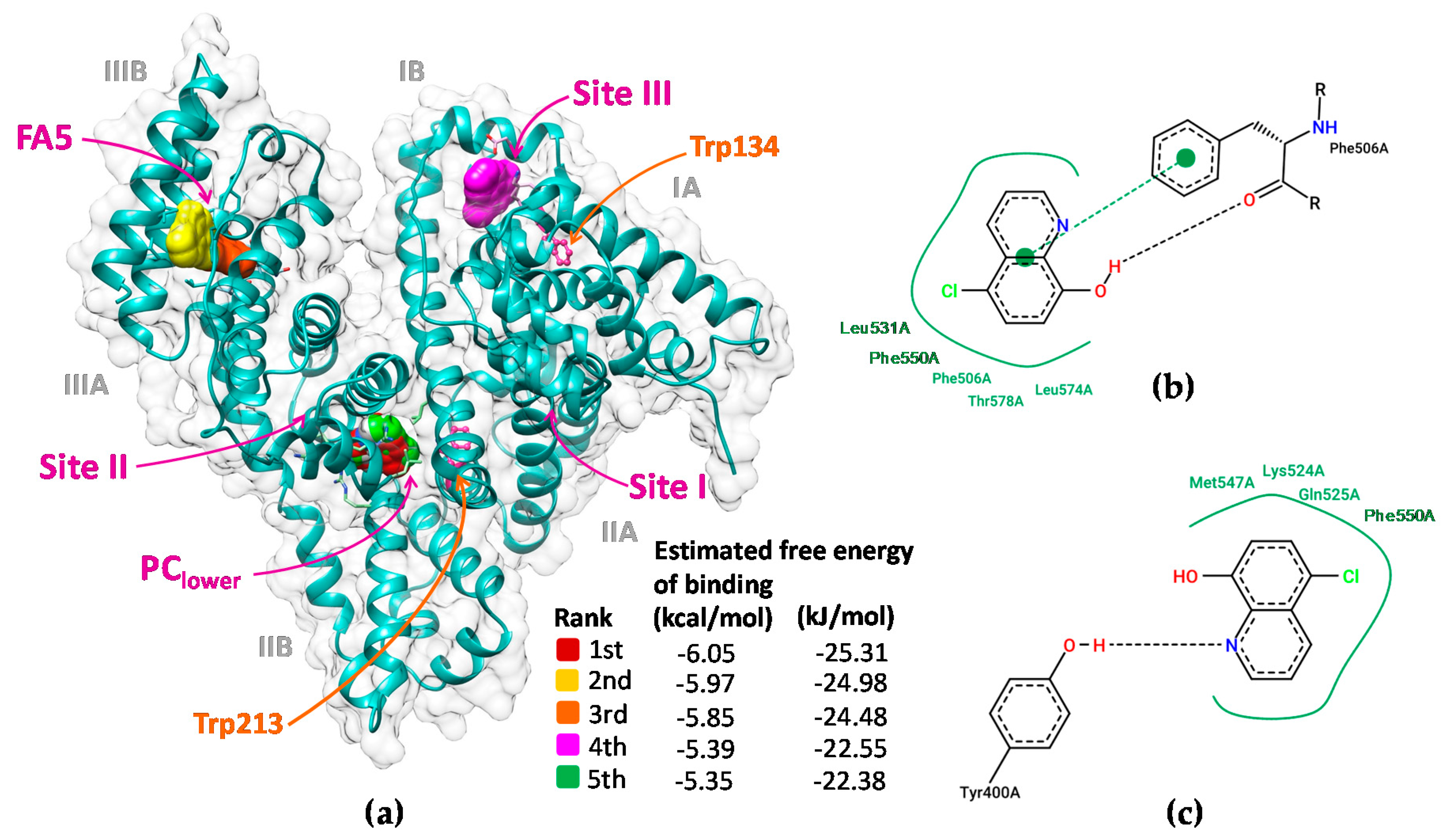
2.8. Molecular Docking for Elucidating the Major Binding Sites of Cloxyquin on BSA
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Steady-State Fluorescence Measurement
3.3. Absorption Measurement
3.4. Site Marker Competitive Analysis
3.5. Dynamic Light Scattering (DLS) experiments
3.6. Circular Dichroism (CD) Spectrometry
3.7. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SA | Serum Albumin |
| BSA | Bovine Serum Albumin |
| HSA | Human Serum Albumin |
| MIC | Minimum Inhibition Concentration |
| TRESK | TWIK-related spinal cord potassium channel |
| FA3 | Fatty acid binding site 3 |
| FA5 | Fatty acid binding site 5 |
| PClower | Protein cleft between subdomains IIA and IIIA |
| K | Kelvin |
| UV-Vis | Ultraviolet and Visible light |
| CD | Circular Dichroism spectroscopy |
| FRET | Fluorescence Resonance Energy Transfer |
| DLS | Dynamic Light Scattering |
| Arg | Arginine |
| GluLeu | GlutamicLeucine |
| Phe | Phenylalanine |
| Ser | Serine |
| Trp | Tryptophan |
| Tyr | Tyrosine |
References
- Cherdtrakulkiat, R.; Worachartcheewan, A.; Tantimavanich, S.; Lawung, R.; Sinthupoom, N.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Discovery of novel halogenated 8-hydroxyquinoline-based anti-MRSA agents: In vitro and QSAR studies. Drug Dev. Res. 2019, 1–9. [Google Scholar] [CrossRef]
- Cherdtrakulkiat, R.; Boonpangrak, S.; Sinthupoom, N.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Derivatives (halogen, nitro and amino) of 8-hydroxyquinoline with highly potent antimicrobial and antioxidant activities. Biochem. Biophys. Rep. 2016, 6, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Hongmanee, P.; Rukseree, K.; Buabut, B.; Somsri, B.; Palittapongarnpim, P. In vitro activities of cloxyquin (5-chloroquinolin-8-ol) against Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007, 51, 1105–1106. [Google Scholar] [CrossRef] [Green Version]
- Fritzler, J.M.; Zhu, G. Novel anti-Cryptosporidium activity of known drugs identified by high-throughput screening against parasite fatty acyl-CoA binding protein (ACBP). J. Antimicrob. Chemother. 2011, 67, 609–617. [Google Scholar] [CrossRef]
- Gershon, H.; Gershon, M.; Clarke, D.D. Synergistic mixtures of fungitoxic monochloro and dichloro-8-quinolinols against five fungi. Mycopathologia 2004, 158, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Nadtochiy, S.M.; Urciuoli, W.R.; Brookes, P.S. The cardioprotective compound cloxyquin uncouples mitochondria and induces autophagy. Am. J. Physiol. Heart Circ. Physiol. 2016, 310, H29–H38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lengyel, M.; Dobolyi, A.; Czirjak, G.; Enyedi, P. Selective and state-dependent activation of TRESK (K2P 18.1) background potassium channel by cloxyquin. Br. J. Pharmacol. 2017, 174, 2102–2113. [Google Scholar] [CrossRef] [Green Version]
- Wright, P.D.; Weir, G.; Cartland, J.; Tickle, D.; Kettleborough, C.; Cader, M.Z.; Jerman, J. Cloxyquin (5-chloroquinolin-8-ol) is an activator of the two-pore domain potassium channel TRESK. Biochem. Biophys. Res. Commun. 2013, 441, 463–468. [Google Scholar] [CrossRef]
- Van Hau, T.; Ruankham, W.; Suwanjang, W.; Songtawee, N.; Wongchitrat, P.; Pingaew, R.; Prachayasittikul, V.; Prachayasittikul, S.; Phopin, K. Repurposing of nitroxoline drug for the prevention of neurodegeneration. Chem. Res. Toxicol. 2019, 2182–2191. [Google Scholar] [CrossRef]
- Ma, Z.; Moulton, B. A novel polymorph of 5-chloro-8-hydroxyquinoline with improved water solubility and faster dissolution rate. J. Chem. Crystallogr. 2009, 39, 913–918. [Google Scholar] [CrossRef]
- Kolehmainen, E.; Salo, H.; Korpela, J. Gelation behavior of 5-chloro-8-hydroxyquinoline, an antituberculosis agent in aqueous alcohol solutions. Antibiotics 2012, 1, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Makarska-Bialokoz, M. Investigation of the binding affinity in vitamin B12—Bovine serum albumin system using various spectroscopic methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 184, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Makarska-Bialokoz, M. Interactions of hemin with bovine serum albumin and human hemoglobin: A fluorescence quenching study. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 193, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagalkoti, J.T.; Joshi, S.D.; Nandibewoor, S.T. Spectral and molecular modelling studies of sulfadoxine interaction with bovine serum albumin. J. Photochem. Photobiol. A 2019, 382, 111871. [Google Scholar] [CrossRef]
- Moriyama, Y.; Ohta, D.; Hachiya, K.; Mitsui, Y.; Takeda, K. Fluorescence behavior of tryptophan residues of bovine and human serum albumins in ionic surfactant solutions: A comparative study of the two and one tryptophan(s) of bovine and human albumins. J. Protein Chem. 1996, 15, 265–272. [Google Scholar] [CrossRef]
- Agudelo, D.; Bourassa, P.; Bruneau, J.; Berube, G.; Asselin, E.; Tajmir-Riahi, H.A. Probing the binding sites of antibiotic drugs doxorubicin and N-(trifluoroacetyl) doxorubicin with human and bovine serum albumins. PLoS ONE 2012, 7, e43814. [Google Scholar] [CrossRef] [Green Version]
- Evoli, S.; Mobley, D.L.; Guzzi, R.; Rizzuti, B. Multiple binding modes of ibuprofen in human serum albumin identified by absolute binding free energy calculations. Phys. Chem. Chem. Phys. 2016, 18, 32358–32368. [Google Scholar] [CrossRef] [Green Version]
- Abou-Zied, O.K.; Al-Lawatia, N.; Elstner, M.; Steinbrecher, T.B. Binding of hydroxyquinoline probes to human serum albumin: Combining molecular modeling and Förster’s resonance energy transfer spectroscopy to understand flexible ligand binding. J. Phys. Chem. B 2013, 117, 1062–1074. [Google Scholar] [CrossRef]
- Zhang, W.; Xiong, X.; Wang, F.; Li, L.; Zhang, Y.; Xiao, W.; Liu, Y. Investigation of nitroxoline-human serum albumin interactions by spectroscopic methods. Sci. China Chem. 2014, 57, 1690–1695. [Google Scholar] [CrossRef]
- Shiriskar, S.M.; Agarwal, N.; Pissurlenkar, R.R.S.; Ahmad, B. Effects of 2-amino-8-hydroxyquinoline interaction on the conformation of physiological isomers of human serum albumin. Eur. Biophys. J. 2015, 44, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Tantimongcolwat, T.; Prachayasittikul, S.; Prachayasittikul, V. Unravelling the interaction mechanism between clioquinol and bovine serum albumin by multi-spectroscopic and molecular docking approaches. Spectrochim. Acta Part A 2019, 216, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Ghisaidoobe, A.B.; Chung, S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: A focus on Forster resonance energy transfer techniques. Int. J. Mol. Sci. 2014, 15, 22518–22538. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Kluwer Academic/Plenum: New York, NY, USA, 2006. [Google Scholar]
- Liu, J.; He, Y.; Liu, D.; He, Y.; Tang, Z.; Lou, H.; Huo, Y.; Cao, X. Characterizing the binding interaction of astilbin with bovine serum albumin: A spectroscopic study in combination with molecular docking technology. RSC Adv. 2018, 8, 7280–7286. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Chen, L.; Ma, L.; Liu, S.; Zhou, H.; Zhang, Y.; Guo, T.; Liu, W.; Dai, H.; Yu, Y. Fluorescence spectroscopic investigation of competitive interactions between quercetin and aflatoxin B (1) for binding to human serum albumin. Toxins (Basel) 2019, 11, 214. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Yao, N.; Xu, H.; Wang, T.; Li, G.; Li, Z. Characterization of the interaction between eupatorin and bovine serum albumin by spectroscopic and molecular modeling methods. Int. J Mol. Sci. 2013, 14, 14185–14203. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Yang, J.; Wu, X.; Huang, X.; Wang, F.; Liu, S. Unfolding and refolding of bovine serum albumin induced by cetylpyridinium bromide. Biophys. J. 2005, 88, 3518–3524. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-Y.; Huang, H.-M.; Lin, C.-C.; Lin, F.-Y.; Chan, Y.-C. Effect of temperature on hydrophobic interaction between proteins and hydrophobic adsorbents: Studies by isothermal titration calorimetry and the van’t Hoff equation. Langmuir 2003, 19, 9395–9403. [Google Scholar] [CrossRef]
- Wang, R.; Liu, Y.; Hu, X.; Pan, J.; Gong, D.; Zhang, G. New insights into the binding mechanism between osthole and β-lactoglobulin: Spectroscopic, chemometrics and docking studies. Food Res. Int. 2019, 120, 226–234. [Google Scholar] [CrossRef]
- Wani, T.A.; Bakheit, A.H.; Zargar, S.; Bhat, M.A.; Al-Majed, A.A. Molecular docking and experimental investigation of new indole derivative cyclooxygenase inhibitor to probe its binding mechanism with bovine serum albumin. Bioorg. Chem. 2019, 89, 103010. [Google Scholar] [CrossRef]
- Patil, S.R.; Salunkhe, S.M.; Wakshe, S.B.; Karnik, K.S.; Sarkate, A.P.; Patrawale, A.A.; Anbhule, P.V.; Kolekar, G.B. Spectral elucidation with molecular docking study between isatin analogous and bovine serum albumin. Chem. Data Collect. 2019, 22, 100254. [Google Scholar] [CrossRef]
- Cao, X.; He, Y.; Liu, D.; He, Y.; Hou, X.; Cheng, Y.; Liu, J. Characterization of interaction between scoparone and bovine serum albumin: Spectroscopic and molecular docking methods. RSC Adv. 2018, 8, 25519–25525. [Google Scholar] [CrossRef] [Green Version]
- Suryawanshi, V.D.; Walekar, L.S.; Gore, A.H.; Anbhule, P.V.; Kolekar, G.B. Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin. J. Pharm. Anal. 2016, 6, 56–63. [Google Scholar] [CrossRef] [Green Version]
- Makarska-Bialokoz, M.; Lipke, A. Study of the binding interactions between uric acid and bovine serum albumin using multiple spectroscopic techniques. J. Mol. Liq. 2019, 276, 595–604. [Google Scholar] [CrossRef]
- Katrahalli, U.; Chanabasappa Yallur, B.; Manjunatha, D.H.; Krishna, P.M. BSA interaction and DNA cleavage studies of anti-bacterial benzothiazol-2-yl-malonaldehyde. J. Mol. Struct. 2019, 1196, 96–104. [Google Scholar] [CrossRef]
- Khatun, S.; Rabbani, G. A comparative biophysical and in-silico studies on the interactions of ticlopidine hydrochloride with two serum albumins. J. Chem. Thermodyn. 2019, 131, 9–20. [Google Scholar] [CrossRef]
- Shalaeva, Y.V.; Morozova, J.E.; Shumatbaeva, A.M.; Nizameev, I.R.; Kadirov, M.K.; Antipin, I.S. Binding of l-tryptophan and bovine serum albumin by novel gold nanoparticles capped with amphiphilic sulfonatomethylated calixresorcinarenes. J. Mol. Liq. 2019, 286, 110879. [Google Scholar] [CrossRef]
- Yohannes, G.; Wiedmer, S.K.; Elomaa, M.; Jussila, M.; Aseyev, V.; Riekkola, M.-L. Thermal aggregation of bovine serum albumin studied by asymmetrical flow field-flow fractionation. Anal. Chim. Acta 2010, 675, 191–198. [Google Scholar] [CrossRef]
- Jachimska, B.; Wasilewska, M.; Adamczyk, Z. Characterization of globular protein solutions by dynamic light scattering, electrophoretic mobility, and viscosity measurements. Langmuir 2008, 24, 6866–6872. [Google Scholar] [CrossRef]
- Ritchie, C.W.; Bush, A.I.; Mackinnon, A.; Macfarlane, S.; Mastwyk, M.; MacGregor, L.; Kiers, L.; Cherny, R.; Li, Q.-X.; Tammer, A.; et al. Metal-protein attenuation with iodochlorhydroxyquin (clioquinol) targeting Aβ amyloid deposition and toxicity in alzheimer disease: A pilot phase 2 clinical trial. Arc. Neurol. 2003, 60, 1685–1691. [Google Scholar] [CrossRef]
- Yallur, B.C.; Katrahalli, U.; Krishna, P.M.; Hadagali, M.D. BSA binding and antibacterial studies of newly synthesized 5,6-dihydroimidazo[2,1-b]thiazole-2-carbaldehyde. Spectrochim. Acta Part A 2019, 222, 117192. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, V.; Rajamanikandan, R.; Ilanchelian, M.; Moodley, K.G.; Redhi, G.G. Elucidation of interactions of BSA with [EPMpyr]+[Cl]− using spectroscopic techniques with reference to theoretical thermodynamic and molecular docking studies. J. Mol. Liq. 2019, 273, 634–644. [Google Scholar] [CrossRef]
- Manea, Y.K.; Khan, A.M.T.; Qashqoosh, M.T.A.; Wani, A.A.; Shahadat, M. Ciprofloxacin-supported chitosan/polyphosphate nanocomposite to bind bovine serum albumin: Its application in drug delivery. J. Mol. Liq. 2019, 292, 111337. [Google Scholar] [CrossRef]
- Senthilkumar, R.; Marimuthu, P.; Paul, P.; Manojkumar, Y.; Arunachalam, S.; Eriksson, J.E.; Johnson, M.S. Plasma protein binding of anisomelic acid: Spectroscopy and molecular dynamic simulations. J. Chem. Inf. Modell. 2016, 56, 2401–2412. [Google Scholar] [CrossRef]
- Hazai, E.; Kovács, S.; Demko, L.; Bikadi, Z. DockingServer: Molecular docking calculations online. Acta Pharm. Hung. 2009, 79, 17–21. [Google Scholar]











| T (K) | r2 | ||
|---|---|---|---|
| 290 | 6.373 ± 0.12 | 6.373 ± 0.12 | 0.9978 |
| 300 | 5.415 ± 0.08 | 5.415 ± 0.08 | 0.9988 |
| 310 | 4.566 ± 0.06 | 4.566 ± 0.06 | 0.9990 |
| T (K) | [Cloxyquin] 1 | r2 | |||||
|---|---|---|---|---|---|---|---|
| 290 | High | 0.7870 ± 0.03 | 0.9567 ± 0.01 | 0.9965 | −21.70 ± 0.3 | ||
| Low | 3.1419 ± 0.09 | 0.6402 ± 0.09 | 0.9922 | −52.73 ± 0.7 | |||
| 300 | High | 0.6353 ± 0.05 | 0.9615 ± 0.02 | 0.9939 | 21.6 ± 1.5 | 0.4 ± 0.3 | −21.70 ± 0.4 |
| Low | 1.4231 ± 0.06 | 0.6992 ± 0.07 | 0.9990 | 38.8 ± 0.9 | 48.2 ± 0.5 | −53.21 ± 0.9 | |
| 310 | High | 0.4408 ± 0.07 | 1.0016 ± 0.02 | 0.9942 | −21.71 ± 0.5 | ||
| Low | 1.1204 ± 0.01 | 0.6425 ± 0.01 | 0.9988 | −53.69 ± 1.1 |
| T (K) | n | r2 | ||||
|---|---|---|---|---|---|---|
| 290 | 0.5506 ± 0.01 | 0.755 ± 0.02 | 0.9915 | −21.0 ± 0.2 | ||
| 300 | 1.3590 ± 0.01 | 0.861 ± 0.01 | 0.9975 | 43.4 ± 2.9 | 222.0 ± 10.9 | −23.2 ± 0.3 |
| 310 | 1.7459 ± 0.01 | 0.907 ± 0.02 | 0.9954 | −25.5 ± 0.4 |
| Site Marker | n | r2 | |
|---|---|---|---|
| Without | 1.746 ± 0.01 | 0.91 ± 0.02 | 0.9954 |
| Warfarin | 2.454 ± 0.01 | 0.94 ± 0.02 | 0.9943 |
| Ibuprofen | 1.424 ± 0.01 | 0.89 ± 0.02 | 0.9923 |
| Digitoxin | 1.239 ± 0.01 | 0.88 ± 0.04 | 0.9828 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phopin, K.; Ruankham, W.; Prachayasittikul, S.; Prachayasittikul, V.; Tantimongcolwat, T. Insight into the Molecular Interaction of Cloxyquin (5-chloro-8-hydroxyquinoline) with Bovine Serum Albumin: Biophysical Analysis and Computational Simulation. Int. J. Mol. Sci. 2020, 21, 249. https://doi.org/10.3390/ijms21010249
Phopin K, Ruankham W, Prachayasittikul S, Prachayasittikul V, Tantimongcolwat T. Insight into the Molecular Interaction of Cloxyquin (5-chloro-8-hydroxyquinoline) with Bovine Serum Albumin: Biophysical Analysis and Computational Simulation. International Journal of Molecular Sciences. 2020; 21(1):249. https://doi.org/10.3390/ijms21010249
Chicago/Turabian StylePhopin, Kamonrat, Waralee Ruankham, Supaluk Prachayasittikul, Virapong Prachayasittikul, and Tanawut Tantimongcolwat. 2020. "Insight into the Molecular Interaction of Cloxyquin (5-chloro-8-hydroxyquinoline) with Bovine Serum Albumin: Biophysical Analysis and Computational Simulation" International Journal of Molecular Sciences 21, no. 1: 249. https://doi.org/10.3390/ijms21010249







