Conformational Plasticity of the Active Site Entrance in E. coli Aspartate Transcarbamoylase and Its Implication in Feedback Regulation
Abstract
:1. Introduction
2. Results
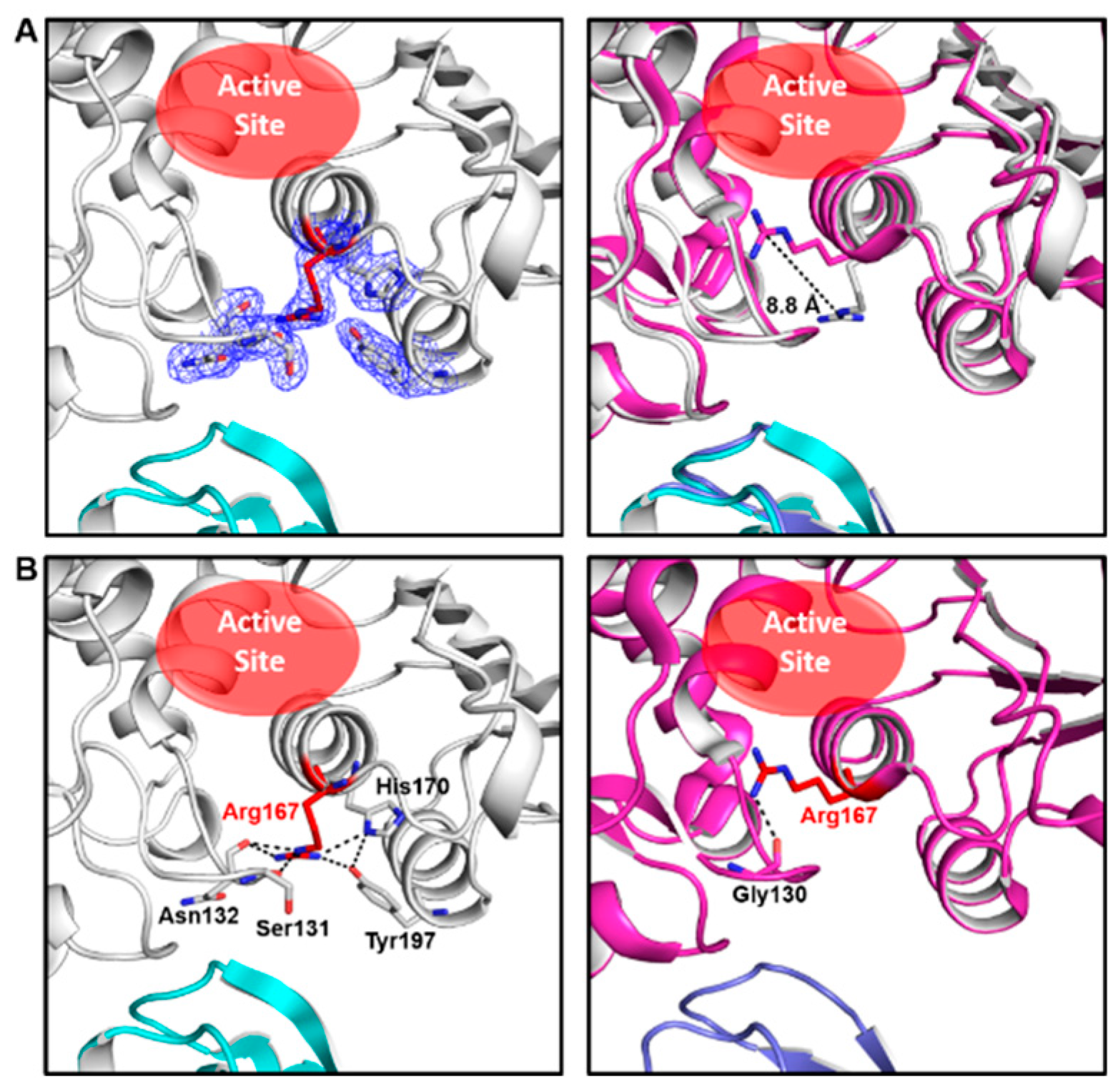
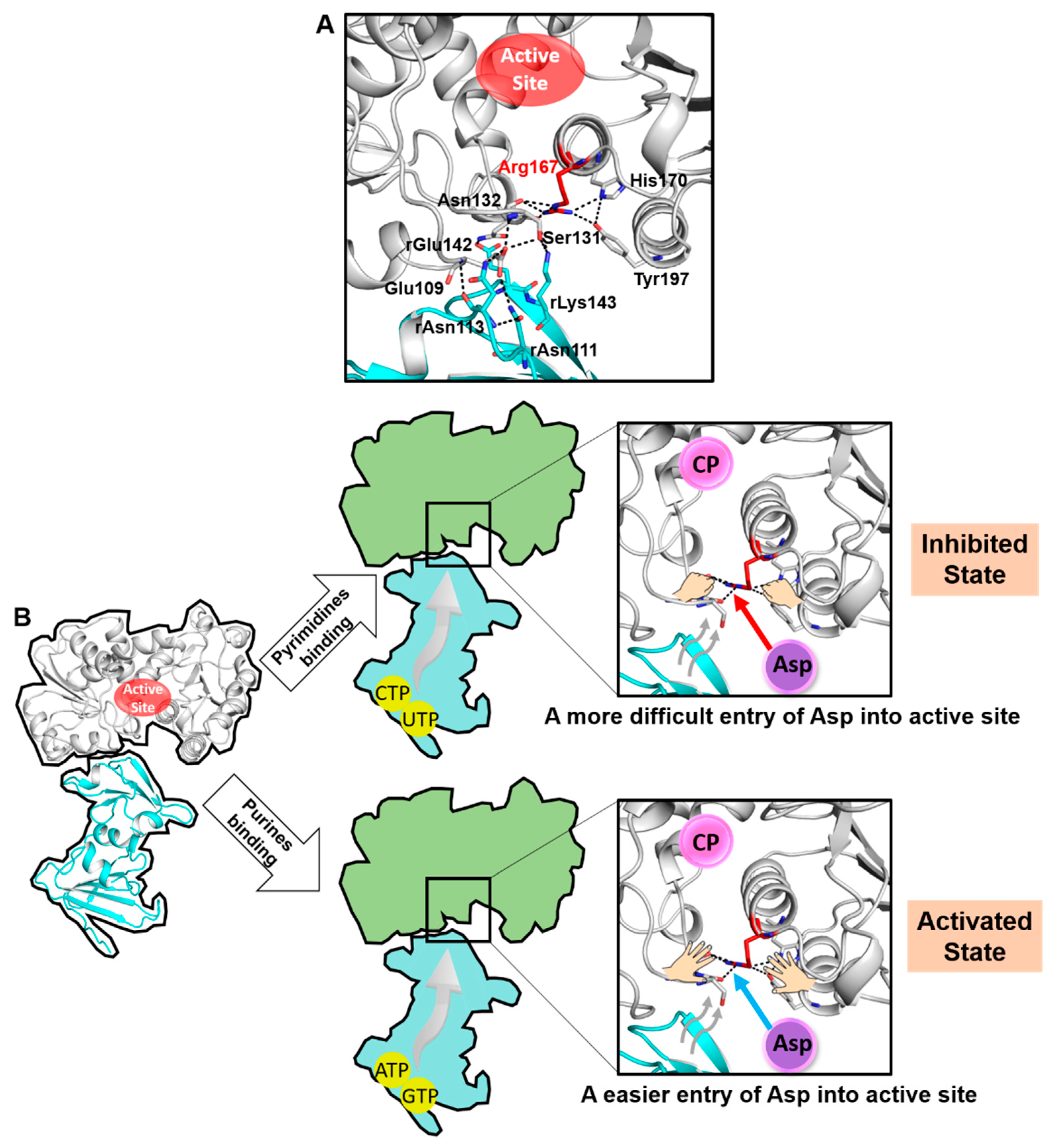
2.1. The Structure of ATCase Holoenzyme with an Uncommon Open Conformation of Arg167 in Its Catalytic Subunit
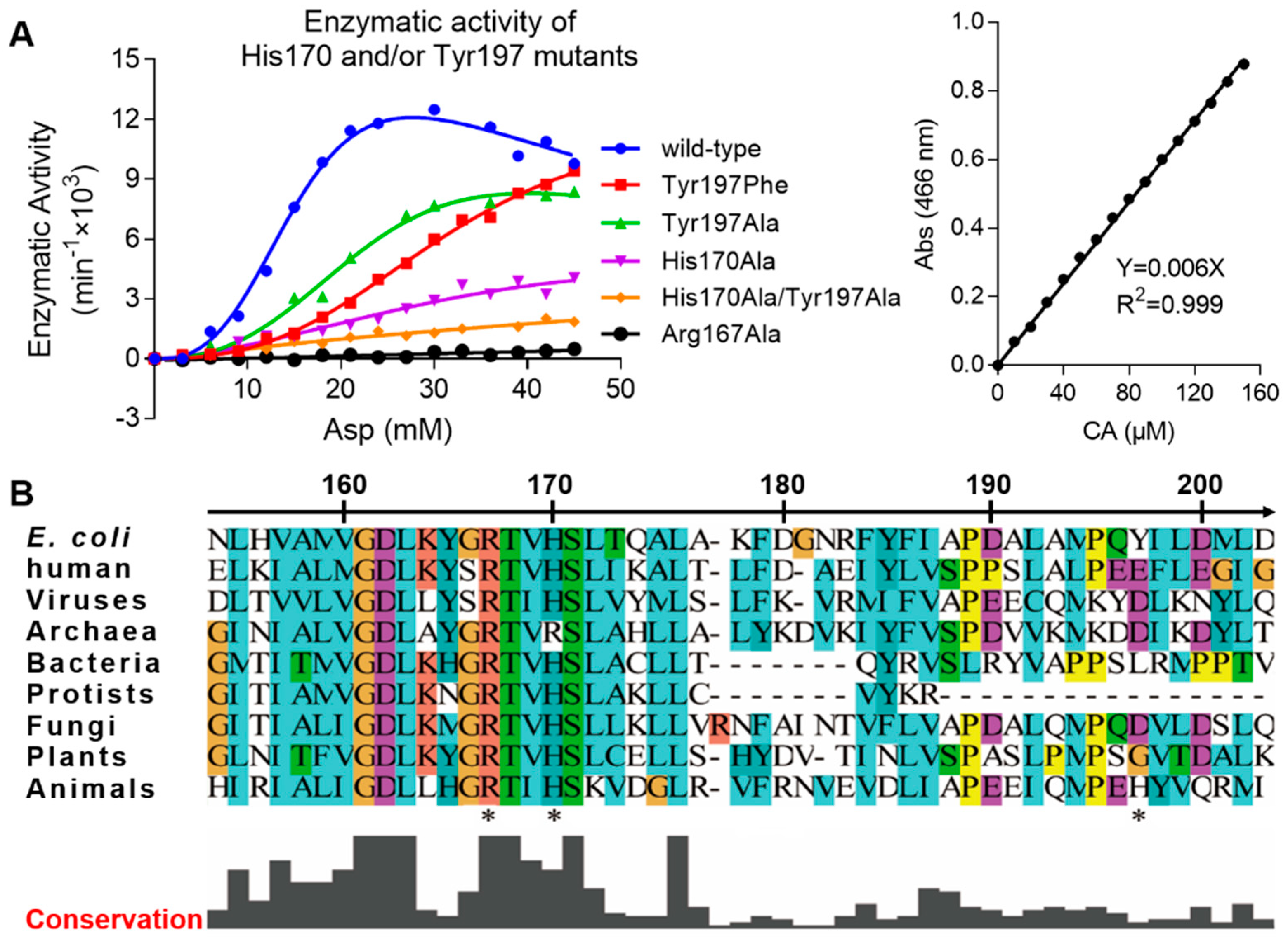
2.2. Enzymatic Activity Assay of ATCase with Mutations of His170 and/or Tyr197
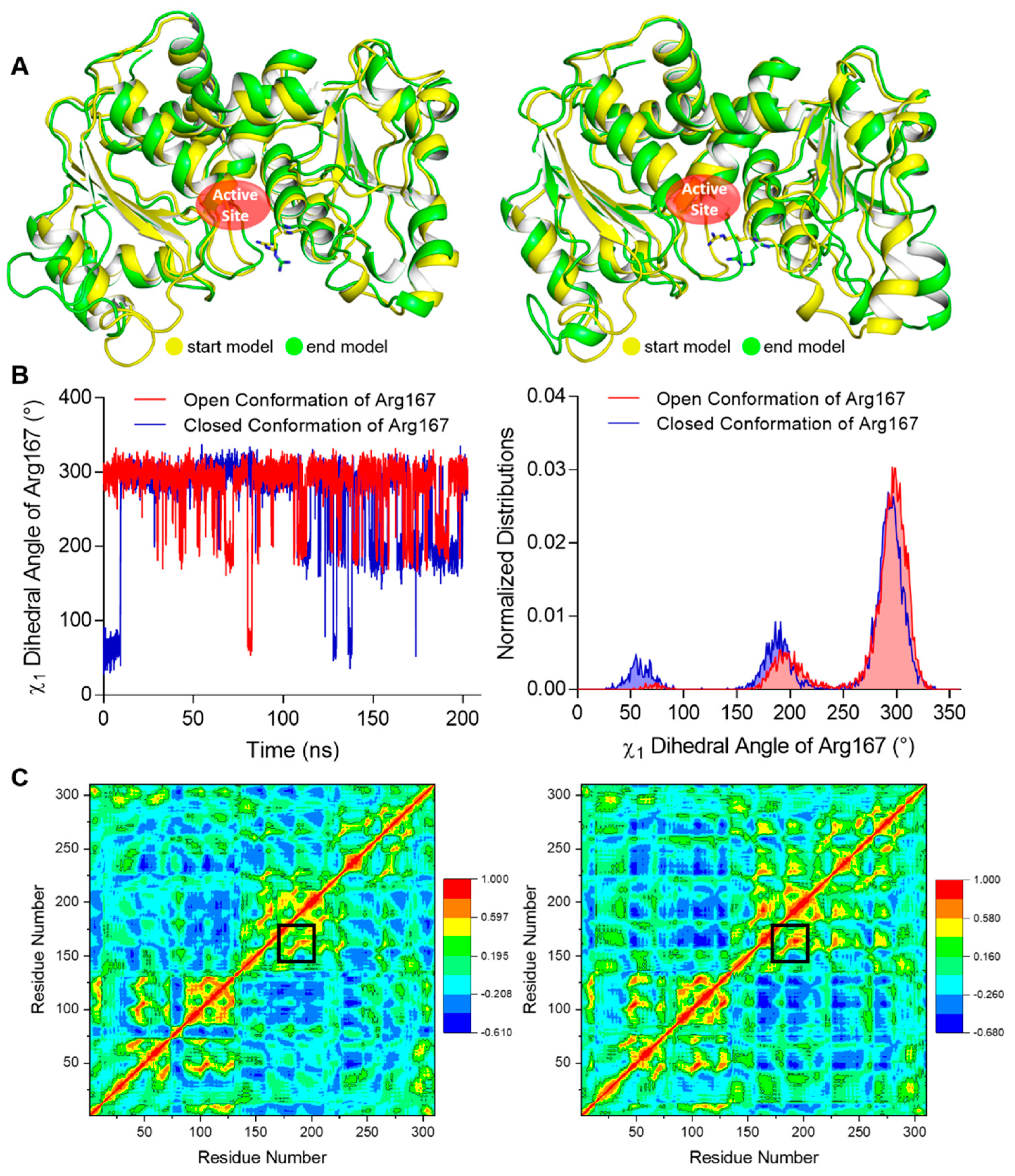
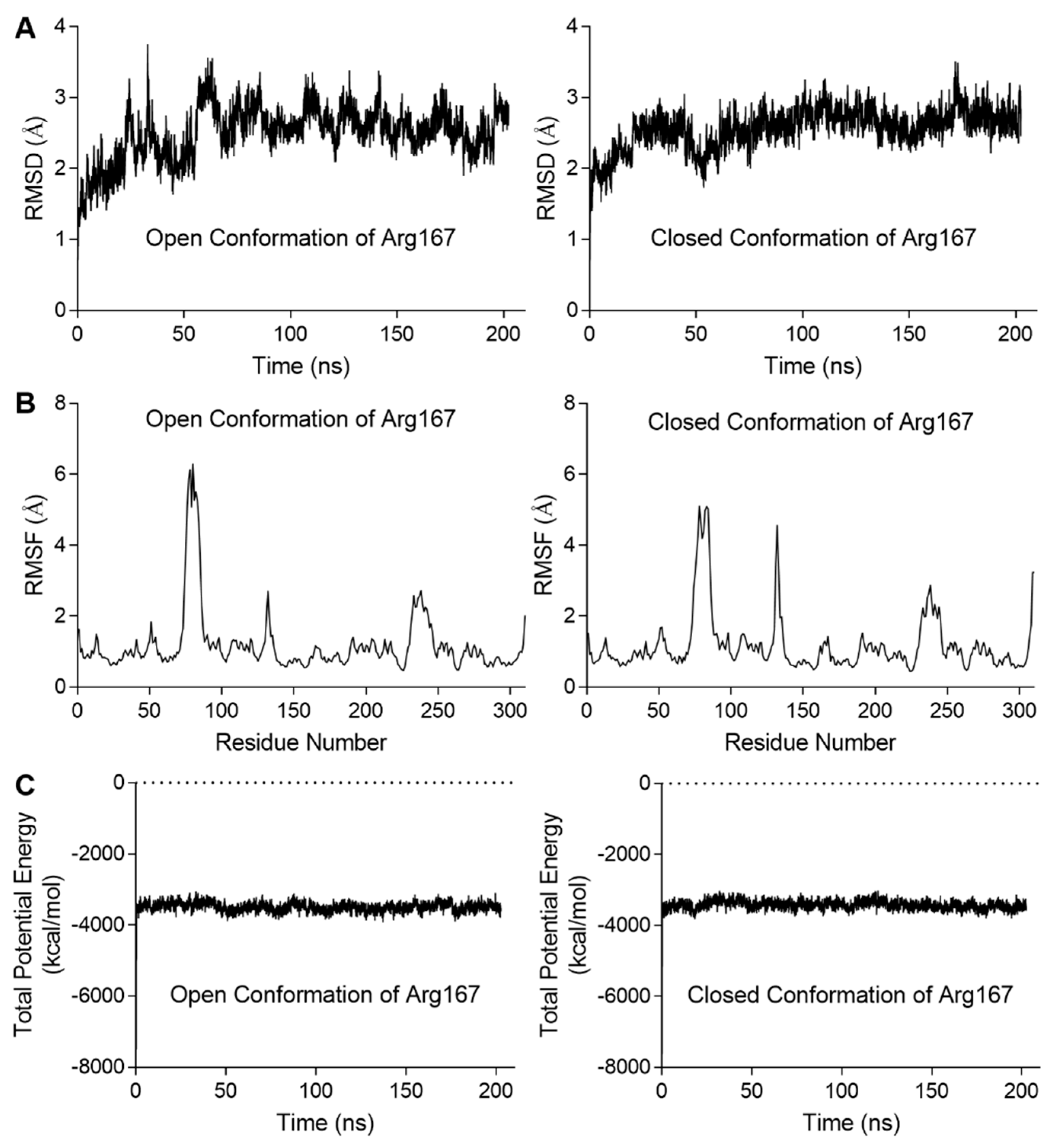
2.3. Molecular Dynamics Simulation for the Stability of the Open and Closed Conformation of Arg167
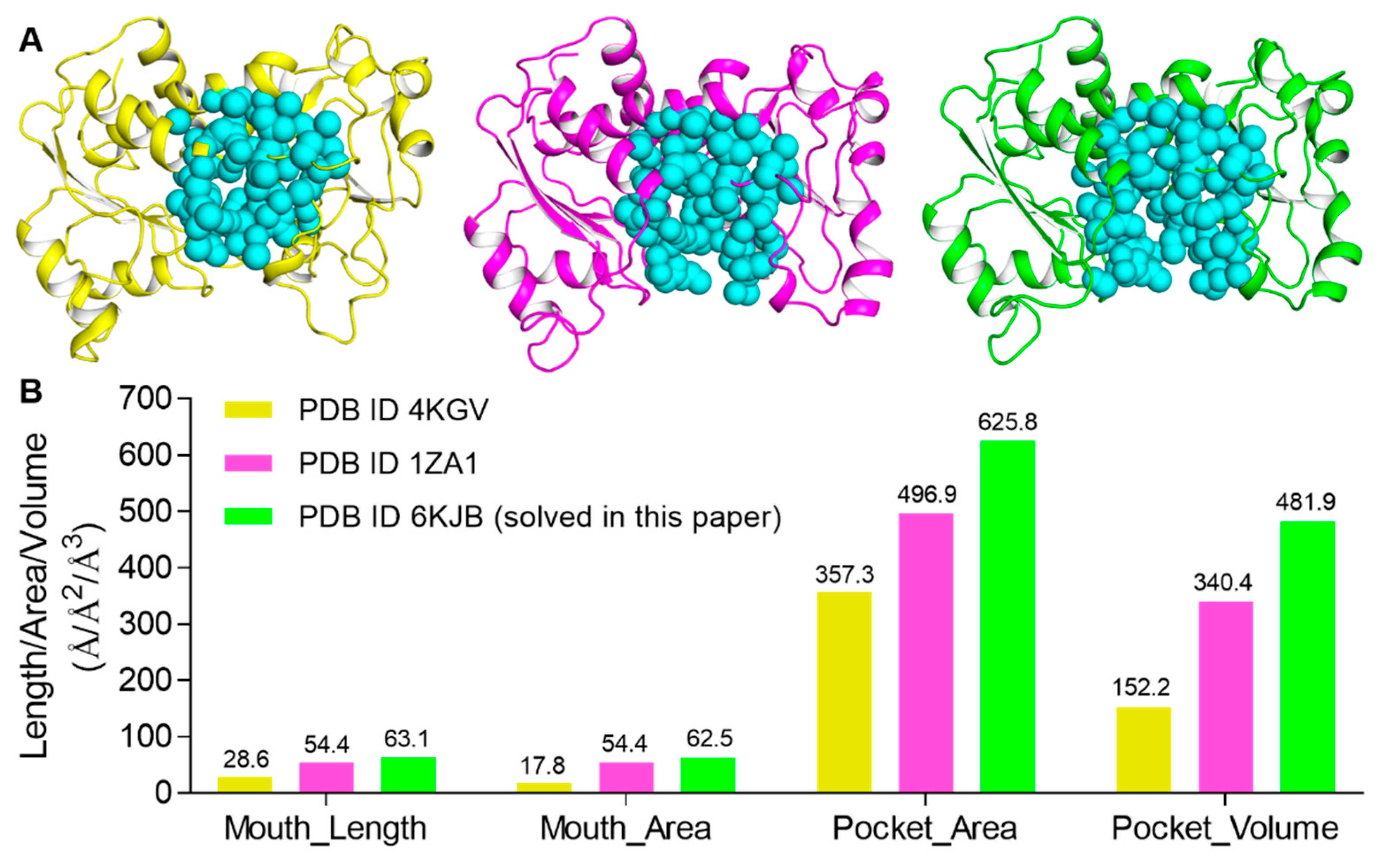
2.4. CASTp Analysis of the Pocket of ATCase with Various Conformations of Arg167
3. Discussion
4. Materials and Methods
4.1. Extractions of ATCase from E. coli
4.2. Crystallization and Structure Determination
4.3. Recombinant Expressions and Purifications of E. coli ATCase and Mutants
4.4. Activity Assay of ATCase and Mutants
4.5. Molecular Dynamic Simulation and CASTp Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lowenstein, J.M.; Cohen, P.P. Studies on the biosynthesis of carbamylaspartic acid. J. Biol. Chem. 1956, 220, 57–70. [Google Scholar]
- Jones, M.E.; Spector, L.; Lipmann, F. Carbamyl Phosphate, The Carbamyl Donor In Enzymatic Citrulline Synthesis1. J. Am. Chem. Soc. 1955, 77, 819–820. [Google Scholar] [CrossRef]
- Reichard, P.; Hanshoff, G. Aspartate Carbamyl Transferase from Escherichia-Coli. Acta Chem. Scand. 1956, 10, 548–566. [Google Scholar] [CrossRef]
- Wedler, F.C. Mechanisms of substrate binding with glutamine synthetase. Equilibrium isotope exchanges with the ovine brain, pea seed, and Escherichia coli enzymes. J. Biol. Chem. 1974, 249, 5080–5087. [Google Scholar]
- Wang, J.; Stieglitz, K.A.; Cardia, J.P.; Kantrowitz, E.R. Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase. Proc. Natl. Acad. Sci. USA 2005, 102, 8881–8886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipscomb, W.N.; Kantrowitz, E.R. Structure and mechanisms of Escherichia coli aspartate transcarbamoylase. Acc. Chem. Res. 2012, 45, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Kantrowitz, E.R. Allostery and cooperativity in Escherichia coli aspartate transcarbamoylase. Arch. Biochem. Biophys. 2012, 519, 81–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howlett, G.J.; Blackburn, M.N.; Compton, J.G.; Schachman, H.K. Allosteric regulation of aspartate transcarbamoylase. Analysis of the structural and functional behavior in terms of a two-state model. Biochemistry 1977, 16, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, J.C.; Pardee, A.B. The enzymology of control by feedback inhibition. J. Biol. Chem. 1962, 237, 891–896. [Google Scholar]
- Fetler, L.; Kantrowitz, E.R.; Vachette, P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc. Natl. Acad. Sci. USA 2007, 104, 495–500. [Google Scholar] [CrossRef] [Green Version]
- Gerhart, J.C.; Pardee, A.B. Aspartate Transcarbamylase, an Enzyme Designed for Feedback Inhibition. Fed. Proc. 1964, 23, 727–735. [Google Scholar] [PubMed]
- Monod, J.; Wyman, J.; Changeux, J.P. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965, 12, 88–118. [Google Scholar] [CrossRef]
- Ladjimi, M.M.; Kantrowitz, E.R. A possible model for the concerted allosteric transition in Escherichia coli aspartate transcarbamylase as deduced from site-directed mutagenesis studies. Biochemistry 1988, 27, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, J.W.; Zhang, Y.; Kantrowitz, E.R. Importance of residues Arg-167 and Gln-231 in both the allosteric and catalytic mechanisms of Escherichia coli aspartate transcarbamoylase. Biochemistry 1990, 29, 3821–3827. [Google Scholar] [CrossRef] [PubMed]
- Newton, C.J.; Kantrowitz, E.R. Importance of domain closure for homotropic cooperativity in Escherichia coli aspartate transcarbamylase. Biochemistry 1990, 29, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.M.; Lipscomb, W.N.; Cho, Y.J.; Honzatko, R.B. Complex of N-phosphonacetyl-l-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J. Mol. Biol. 1988, 204, 725–747. [Google Scholar] [CrossRef]
- Cockrell, G.M.; Zheng, Y.; Guo, W.; Peterson, A.W.; Truong, J.K.; Kantrowitz, E.R. New paradigm for allosteric regulation of Escherichia coli aspartate transcarbamoylase. Biochemistry 2013, 52, 8036–8047. [Google Scholar] [CrossRef]
- Stevens, R.C.; Gouaux, J.E.; Lipscomb, W.N. Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: Crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution. Biochemistry 1990, 29, 7691–7701. [Google Scholar] [CrossRef]
- Lovell, S.C.; Word, J.M.; Richardson, J.S.; Richardson, D.C. The penultimate rotamer library. Proteins 2000, 40, 389–408. [Google Scholar] [CrossRef]
- Ruiz-Ramos, A.; Velazquez-Campoy, A.; Grande-Garcia, A.; Moreno-Morcillo, M.; Ramon-Maiques, S. Structure and Functional Characterization of Human Aspartate Transcarbamoylase, the Target of the Anti-tumoral Drug PALA. Structure 2016, 24, 1081–1094. [Google Scholar] [CrossRef] [Green Version]
- Eisenstein, E.; Markby, D.W.; Schachman, H.K. Changes in stability and allosteric properties of aspartate transcarbamoylase resulting from amino acid substitutions in the zinc-binding domain of the regulatory chains. Proc. Natl. Acad. Sci. USA 1989, 86, 3094–3098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenstein, E.; Markby, D.W.; Schachman, H.K. Heterotropic effectors promote a global conformational change in aspartate transcarbamoylase. Biochemistry 1990, 29, 3724–3731. [Google Scholar] [CrossRef] [PubMed]
- Jancarik, J.; Scott, W.G.; Milligan, D.L.; Koshland, D.E., Jr.; Kim, S.H. Crystallization and preliminary X-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J. Mol. Biol. 1991, 221, 31–34. [Google Scholar] [CrossRef]
- Wang, Q.-S.; Zhang, K.-H.; Cui, Y.; Wang, Z.-J.; Pan, Q.-Y.; Liu, K.; Sun, B.; Zhou, H.; Li, M.-J.; Xu, Q.; et al. Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 2018, 29, 68. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Storoni, L.C.; McCoy, A.J.; Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 432–438. [Google Scholar] [CrossRef]
- Vagin, A.A.; Steiner, R.A.; Lebedev, A.A.; Potterton, L.; McNicholas, S.; Long, F.; Murshudov, G.N. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2184–2195. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Pastra-Landis, S.C.; Foote, J.; Kantrowitz, E.R. An improved colorimetric assay for aspartate and ornithine transcarbamylases. Anal. Biochem. 1981, 118, 358–363. [Google Scholar] [CrossRef]
- Pastra-Landis, S.C.; Evans, D.R.; Lipscomb, W.N. The effect of pH on the cooperative behavior of aspartate transcarbamylase from Escherichia coli. J. Biol. Chem. 1978, 253, 4624–4630. [Google Scholar] [PubMed]
- Case, D.; Betz, R.; Cerutti, D.S.; Cheatham, T.; Darden, T.; Duke, R.; Giese, T.J.; Gohlke, H.; Götz, A.; Homeyer, N.; et al. Amber 2016; University of California: San Francisco, CA, USA, 2016; Available online: http://ambermd.org/doc12/Amber16.pdf (accessed on 31 December 2019). [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, D.J.; Brooks, C.L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004, 121, 10096–10103. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Götz, A.W.; Poole, D.; Le Grand, S.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Item | Value |
|---|---|
| Data collection statistics | |
| Wavelength (Å) | 0.979 |
| Space group | R32 |
| Resolution (Å) | 30.77–2.06 (2.13–2.06) b |
| Rmeas | 0.127 (0.865) |
| Average (I/σ) | 15.8 (3.0) |
| Redundancy | 6.8 (7.3) |
| Completeness (%) | 98.8 (95.8) |
| Unit cell | |
| a, b, c (Å) | 129.7, 129.7, 198.0 |
| α, β, γ (°) | 90, 90, 120 |
| Refinement statistics | |
| Resolution (Å) | 30.77–2.06 |
| Reflections | 39,504 (3808) |
| Rwork/Rfree | 0.18/0.21 |
| Mean B value (Å2) | 47.3 |
| Number of atoms | |
| Protein | 3387 |
| Zinc | 1 |
| Water | 326 |
| RMS (Root-Mean-Square) deviations | |
| Bond lengths (Å) | 0.006 |
| Angles (°) | 1.07 |
| ATCase Type | Vmax (min−1 × 103) | Km (mM) | nH |
|---|---|---|---|
| wild-type ATCase | 13.09 ± 0.26 | 13.84 ± 0.21 | 4.19 ± 0.23 |
| Tyr197Phe mutant | 13.34 ± 1.02 | 32.98 ± 2.02 | 2.76 ± 0.18 |
| Tyr197Ala mutant | 8.80 ± 0.50 | 19.22 ± 0.92 | 3.67 ± 0.70 |
| His170Ala mutant | 7.11 ± 4.06 | 38.76 ± 26.89 | 1.64 ± 0.55 |
| His170Ala&Tyr197Ala mutant | 4.93 ± 4.44 | 66.37 ± 88.54 | 1.17 ± 0.35 |
| Arg167Ala mutant a | - | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, Z.; Wang, N.; Tan, H.; Zheng, J.; Jia, Z. Conformational Plasticity of the Active Site Entrance in E. coli Aspartate Transcarbamoylase and Its Implication in Feedback Regulation. Int. J. Mol. Sci. 2020, 21, 320. https://doi.org/10.3390/ijms21010320
Lei Z, Wang N, Tan H, Zheng J, Jia Z. Conformational Plasticity of the Active Site Entrance in E. coli Aspartate Transcarbamoylase and Its Implication in Feedback Regulation. International Journal of Molecular Sciences. 2020; 21(1):320. https://doi.org/10.3390/ijms21010320
Chicago/Turabian StyleLei, Zhen, Nan Wang, Hongwei Tan, Jimin Zheng, and Zongchao Jia. 2020. "Conformational Plasticity of the Active Site Entrance in E. coli Aspartate Transcarbamoylase and Its Implication in Feedback Regulation" International Journal of Molecular Sciences 21, no. 1: 320. https://doi.org/10.3390/ijms21010320
APA StyleLei, Z., Wang, N., Tan, H., Zheng, J., & Jia, Z. (2020). Conformational Plasticity of the Active Site Entrance in E. coli Aspartate Transcarbamoylase and Its Implication in Feedback Regulation. International Journal of Molecular Sciences, 21(1), 320. https://doi.org/10.3390/ijms21010320






