Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer
Abstract
1. Introduction
2. Results
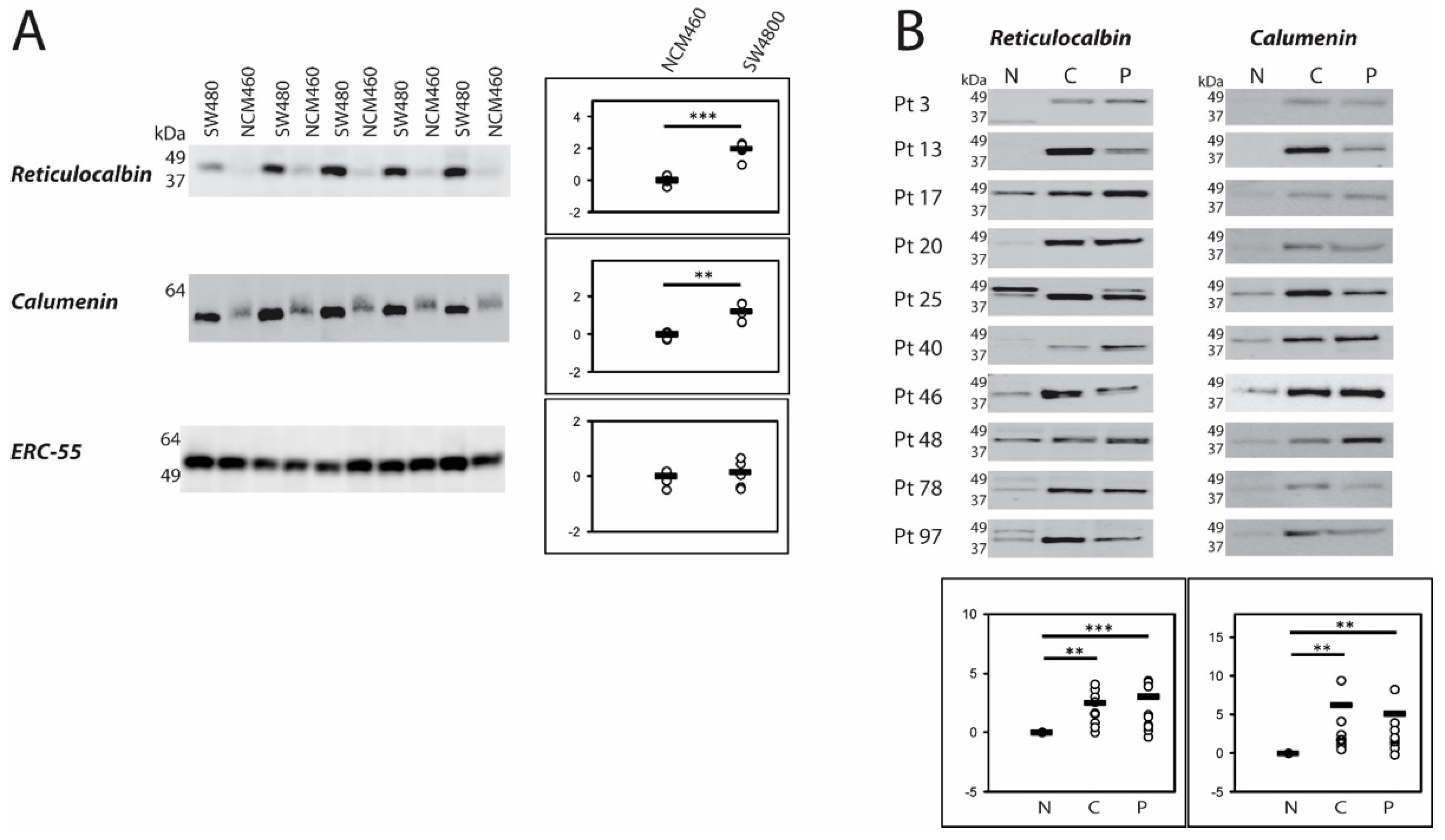
2.1. CREC Protein Expression in NCM460 and SW480 Cell Lines and in Patient Colorectal Cancer (CRC) Tissue
2.2. Top-Down Proteomic Comparison of NCM460 and SW480 Cell Lines
2.3. Bottom-up Proteomic Comparison of NCM460 and HCT 116 Cell Lines
2.4. Immunologic Evaluation of Protein Expression
2.5. S100 Proteins
2.6. Retinol-Binding Protein I
2.7. Protein SET
2.8. Endoplasmin
2.9. Bioinformatic Enrichment Analysis of SW480 versus NCM460
2.10. Bioinformatic Enrichment Analysis of HCT 116 versus NCM460
3. Discussion
3.1. Model Systems Versus Patient Tissue
3.2. Proteomics Methodology
3.3. Comparisons of 2-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) with 1D Western Blotting
3.4. Differentially Expressed Proteins
3.4.1. CREC Proteins
3.4.2. S100 Proteins
3.4.3. Retinol-Binding Protein I
3.4.4. Protein SET
3.4.5. Endoplasmin
3.5. Conclusions
4. Materials and Methods
4.1. Cell Lines
4.2. Tissue Biopsies from Patients with CRC
4.3. Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE)
4.4. Liquid Chromatography–Tandem Mass Spectrometry (LC–MS/MS) for Protein Identification
4.5. Label-Free Quantification Liquid Chromatography–Tandem Mass Spectrometry (LFQ LC–MS/MS)
4.6. Western Blotting
4.7. Bioinformatics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 1D | 1-dimensional |
| 2D-PAGE | 2-dimensional polyacrylamide gel electrophoresis |
| CEA | Carcinoembryonic antigen |
| CRC | Colorectal cancer |
| CREC | Acronym for Cab45, reticulocalbin, ERC-55, calumenin |
| LC-MS/MS | Liquid chromatography - tandem mass spectrometry |
| LFQ | Label-free quantification |
| wt | Wild-type |
References
- Ferlay, J.; Steliarova-Foucher, E.; Lortet-Tieulent, J.; Rosso, S.; Coebergh, J.W.; Comber, H.; Forman, D.; Bray, F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries in 2012. Eur. J. Cancer 2013, 49, 1374–1403. [Google Scholar] [CrossRef]
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef]
- Tsikitis, V.L.; Malireddy, K.; Green, E.A.; Christensen, B.; Whelan, R.; Hyder, J.; Marcello, P.; Larach, S.; Lauter, D.; Sargent, D.J.; et al. Postoperative surveillance recommendations for early stage colon cancer based on results from the clinical outcomes of surgical therapy trial. J. Clin. Oncol. 2009, 27, 3671–3676. [Google Scholar] [CrossRef]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 2011, 6, 479–507. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Gharib, T.G.; Huang, C.C.; Taylor, J.M.; Misek, D.E.; Kardia, S.L.; Giordano, T.J.; Iannettoni, M.D.; Orringer, M.B.; Hanash, S.M.; et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol. Cell Proteom. 2002, 1, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.; Lieberman, D.A.; McFarland, B.; Smith, R.A.; Brooks, D.; Andrews, K.S.; Dash, C.; Giardiello, F.M.; Glick, S.; Levin, T.R.; et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: A joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J. Clin. 2008, 58, 130–160. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Yamashita, K.; Kokuba, Y.; Satoh, T.; Ozawa, H.; Hatate, K.; Ihara, A.; Nakamura, T.; Onozato, W.; Watanabe, M. Diminishing impact of preoperative carcinoembryonic antigen (CEA) in prognosis of Dukes’ C colorectal cancer. Anticancer Res. 2008, 28, 1933–1941. [Google Scholar]
- Yamashita, K.; Watanabe, M. Clinical significance of tumor markers and an emerging perspective on colorectal cancer. Cancer Sci. 2009, 100, 195–199. [Google Scholar] [CrossRef]
- Duffy, M.J.; van Dalen, A.; Haglund, C.; Hansson, L.; Holinski-Feder, E.; Klapdor, R.; Lamerz, R.; Peltomaki, P.; Sturgeon, C.; Topolcan, O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 2007, 43, 1348–1360. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr.; ASCO. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef] [PubMed]
- Motavaf, E.; Sunesen, K.G.; Stender, M.T.; Thorlacius-Ussing, O. Prognostic value of preoperative D-dimer and carcinoembryonic antigen levels in patients undergoing intended curative resection for colorectal cancer: A prospective cohort study. Int. J. Colorectal Dis. 2014, 29, 1427–1432. [Google Scholar] [CrossRef] [PubMed]
- Moyer, M.P.; Manzano, L.A.; Merriman, R.L.; Stauffer, J.S.; Tanzer, L.R. NCM460, a normal human colon mucosal epithelial cell line. In Vitro Cell Dev. Biol. Anim. 1996, 32, 315–317. [Google Scholar] [CrossRef]
- Leibovitz, A.; Stinson, J.C.; McCombs, W.B., 3rd; McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar] [PubMed]
- Brattain, M.G.; Fine, W.D.; Khaled, F.M.; Thompson, J.; Brattain, D.E. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981, 41, 1751–1756. [Google Scholar] [PubMed]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Nimmrich, I.; Erdmann, S.; Melchers, U.; Finke, U.; Hentsch, S.; Moyer, M.P.; Hoffmann, I.; Muller, O. Seven genes that are differentially transcribed in colorectal tumor cell lines. Cancer Lett. 2000, 160, 37–43. [Google Scholar] [CrossRef]
- Honoré, B. The rapidly expanding CREC protein family: Members, localization, function, and role in disease. Bioessays 2009, 31, 262–277. [Google Scholar] [CrossRef]
- Honoré, B.; Vorum, H. The CREC family, a novel family of multiple EF-hand, low-affinity Ca(2+)-binding proteins localised to the secretory pathway of mammalian cells. FEBS Lett. 2000, 466, 11–18. [Google Scholar] [CrossRef]
- Liu, Z.; Brattain, M.G.; Appert, H. Differential display of reticulocalbin in the highly invasive cell line, MDA-MB-435, versus the poorly invasive cell line, MCF-7. Biochem. Biophys. Res. Commun. 1997, 231, 283–289. [Google Scholar] [CrossRef]
- Yu, L.R.; Zeng, R.; Shao, X.X.; Wang, N.; Xu, Y.H.; Xia, Q.C. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis 2000, 21, 3058–3068. [Google Scholar] [CrossRef]
- Hirano, T.; Kato, H.; Maeda, M.; Gong, Y.; Shou, Y.; Nakamura, M.; Maeda, J.; Yashima, K.; Kato, Y.; Akimoto, S.; et al. Identification of postoperative adjuvant chemotherapy responders in non-small cell lung cancer by novel biomarker. Int. J. Cancer 2005, 117, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Pass, H.I.; Liu, Z.; Wali, A.; Bueno, R.; Land, S.; Lott, D.; Siddiq, F.; Lonardo, F.; Carbone, M.; Draghici, S. Gene expression profiles predict survival and progression of pleural mesothelioma. Clin. Cancer Res. 2004, 10, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Hui, Z.; Wang, D.; Jiang, G.; Wang, J.; Zhang, G. Molecular cloning, identification and analysis of lung squamous cell carcinoma-related genes. Lung Cancer 2002, 38, 235–241. [Google Scholar] [CrossRef]
- Cavallo, F.; Astolfi, A.; Iezzi, M.; Cordero, F.; Lollini, P.L.; Forni, G.; Calogero, R. An integrated approach of immunogenomics and bioinformatics to identify new Tumor Associated Antigens (TAA) for mammary cancer immunological prevention. BMC Bioinform. 2005, 6, S7. [Google Scholar] [CrossRef][Green Version]
- Vazquez-Ortiz, G.; Garcia, J.A.; Ciudad, C.J.; Noe, V.; Penuelas, S.; Lopez-Romero, R.; Mendoza-Lorenzo, P.; Pina-Sanchez, P.; Salcedo, M. Differentially expressed genes between high-risk human papillomavirus types in human cervical cancer cells. Int. J. Gynecol. Cancer 2007, 17, 484–491. [Google Scholar] [CrossRef]
- Strefford, J.C.; Lane, T.M.; Hill, A.; LeRoux, L.; Foot, N.J.; Shipley, J.; Oliver, R.T.; Lu, Y.J.; Young, B.D. Molecular characterisation of the t(1; 15)(p22; q22) translocation in the prostate cancer cell line LNCaP. Cytogenet. Genome Res. 2006, 112, 45–52. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- De Wit, M.; Fijneman, R.J.; Verheul, H.M.; Meijer, G.A.; Jimenez, C.R. Proteomics in colorectal cancer translational research: Biomarker discovery for clinical applications. Clin. Biochem. 2013, 46, 466–479. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L.; The Consortium for Top Down Proteomics. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Ostasiewicz, P.; Dus, K.; Zielinska, D.F.; Gnad, F.; Mann, M. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Syst. Biol. 2012, 8, 611. [Google Scholar] [CrossRef] [PubMed]
- Tanca, A.; Addis, M.F.; Simula, M.P.; Pagnozzi, D.; Biosa, G.; Pisanu, S.; Garziera, M.; Cannizzaro, R.; Canzonieri, V.; de Re, V.; et al. Evaluation of the suitability of archival Bouin-fixed paraffin-embedded tissue specimens to proteomic investigation. Electrophoresis 2012, 33, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Yabe, D.; Nakamura, T.; Kanazawa, N.; Tashiro, K.; Honjo, T. Calumenin, a Ca2+-binding protein retained in the endoplasmic reticulum with a novel carboxyl-terminal sequence, HDEF. J. Biol. Chem. 1997, 272, 18232–18239. [Google Scholar] [CrossRef] [PubMed]
- Vorum, H.; Hager, H.; Christensen, B.M.; Nielsen, S.; Honoré, B. Human calumenin localizes to the secretory pathway and is secreted to the medium. Exp. Cell Res. 1999, 248, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Muramatsu, T. Reticulocalbin, a novel endoplasmic reticulum resident Ca(2+)-binding protein with multiple EF-hand motifs and a carboxyl-terminal HDEL sequence. J. Biol. Chem. 1993, 268, 699–705. [Google Scholar]
- Komatsu, K.; Kobune-Fujiwara, Y.; Andoh, A.; Ishiguro, S.; Hunai, H.; Suzuki, N.; Kameyama, M.; Murata, K.; Miyoshi, J.; Akedo, H.; et al. Increased expression of S100A6 at the invading fronts of the primary lesion and liver metastasis in patients with colorectal adenocarcinoma. Br. J. Cancer 2000, 83, 769–774. [Google Scholar] [CrossRef]
- Bronckart, Y.; Decaestecker, C.; Nagy, N.; Harper, L.; Schafer, B.W.; Salmon, I.; Pochet, R.; Kiss, R.; Heizman, C.W. Development and progression of malignancy in human colon tissues are correlated with expression of specific Ca(2+)-binding S100 proteins. Histol. Histopathol. 2001, 16, 707–712. [Google Scholar]
- Leclerc, E.; Heizmann, C.W. The importance of Ca2+/Zn2+ signaling S100 proteins and RAGE in translational medicine. Front. Biosci. 2011, 3, 1232–1262. [Google Scholar]
- Komatsu, K.; Murata, K.; Kameyama, M.; Ayaki, M.; Mukai, M.; Ishiguro, S.; Miyoshi, J.; Tatsuta, M.; Inoue, M.; Nakamura, H. Expression of S100A6 and S100A4 in matched samples of human colorectal mucosa, primary colorectal adenocarcinomas and liver metastases. Oncology 2002, 63, 192–200. [Google Scholar] [CrossRef]
- Ghoul, A.; Serova, M.; Astorgues-Xerri, L.; Bieche, I.; Bousquet, G.; Varna, M.; Vidaud, M.; Phillips, E.; Weill, S.; Benhadji, K.A.; et al. Epithelial-to-mesenchymal transition and resistance to ingenol 3-angelate, a novel protein kinase C modulator, in colon cancer cells. Cancer Res. 2009, 69, 4260–4269. [Google Scholar] [CrossRef]
- Taylor, S.; Herrington, S.; Prime, W.; Rudland, P.S.; Barraclough, R. S100A4 (p9Ka) protein in colon carcinoma and liver metastases: Association with carcinoma cells and T-lymphocytes. Br. J. Cancer 2002, 86, 409–416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gongoll, S.; Peters, G.; Mengel, M.; Piso, P.; Klempnauer, J.; Kreipe, H.; von Wasielewski, R. Prognostic significance of calcium-binding protein S100A4 in colorectal cancer. Gastroenterology 2002, 123, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Boye, K.; Maelandsmo, G.M. S100A4 and metastasis: A small actor playing many roles. Am. J. Pathol. 2010, 176, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Basu, T.K.; Chan, U.M.; Fields, A.L.; McPherson, T.A. Retinol and postoperative colorectal cancer patients. Br. J. Cancer 1985, 51, 61–65. [Google Scholar] [CrossRef]
- Ali, N.A.; McKay, M.J.; Molloy, M.P. Proteomics of Smad4 regulated transforming growth factor-beta signalling in colon cancer cells. Mol. Biosyst. 2010, 6, 2332–2338. [Google Scholar] [CrossRef]
- Lou, J.; Fatima, N.; Xiao, Z.; Stauffer, S.; Smythers, G.; Greenwald, P.; Ali, I.U. Proteomic profiling identifies cyclooxygenase-2-independent global proteomic changes by celecoxib in colorectal cancer cells. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1598–1606. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Wang, Q.; Guo, L.; Ying, X.; Zhao, Y. Immunolocalisation of heat shock protein 72 and glycoprotein 96 in colonic adenocarcinoma. Acta Histochem. 2008, 110, 117–123. [Google Scholar] [CrossRef]
- Honoré, B. Proteomic Protocols for Differential Protein Expression Analyses. Methods Mol. Biol. 2020, 2110, 47–58. [Google Scholar] [CrossRef]
- Kamper, P.; Ludvigsen, M.; Bendix, K.; Hamilton-Dutoit, S.; Rabinovich, G.A.; Møller, M.B.; Nyengaard, J.R.; Honoré, B.; d’Amore, F. Proteomic analysis identifies galectin-1 as a predictive biomarker for relapsed/refractory disease in classical Hodgkin lymphoma. Blood 2011, 117, 6638–6649. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Christakopoulos, C.; Cehofski, L.J.; Christensen, S.R.; Vorum, H.; Honoré, B. Proteomics reveals a set of highly enriched proteins in epiretinal membrane compared with inner limiting membrane. Exp. Eye Res. 2019, 186, 107722. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Hu, X.; Du, S.; Yu, J.; Yang, X.; Yang, C.; Zhou, D.; Wang, Q.; Qin, S.; Yan, X.; He, L.; et al. Common housekeeping proteins are upregulated in colorectal adenocarcinoma and hepatocellular carcinoma, making the total protein a better “housekeeper”. Oncotarget 2016, 7, 66679–66688. [Google Scholar] [CrossRef]
- Xu, L.; Luo, H.; Wang, R.; Wu, W.W.; Phue, J.N.; Shen, R.F.; Juhl, H.; Wu, L.; Alterovitz, W.L.; Simonyan, V.; et al. Novel reference genes in colorectal cancer identify a distinct subset of high stage tumors and their associated histologically normal colonic tissues. BMC Med. Genet. 2019, 20, 138. [Google Scholar] [CrossRef]
- Hansen, G.A.; Ludvigsen, M.; Jacobsen, C.; Cangemi, C.; Rasmussen, L.M.; Vorum, H.; Honoré, B. Fibulin-1C, C1 Esterase Inhibitor and Glucose Regulated Protein 75 Interact with the CREC Proteins, Calumenin and Reticulocalbin. PLoS ONE 2015, 10, e0132283. [Google Scholar] [CrossRef][Green Version]
- Ludvigsen, M.; Jacobsen, C.; Maunsbach, A.B.; Honoré, B. Identification and characterization of novel ERC-55 interacting proteins: Evidence for the existence of several ERC-55 splicing variants; including the cytosolic ERC-55-C. Proteomics 2009, 9, 5267–5287. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [PubMed]





| Patient no. | Sex | Age by the Time of Diagnosis | Localization | T | N | M | Rt-Type | Recurrence | Death Due to CRC |
|---|---|---|---|---|---|---|---|---|---|
| 3 | M | 63 | 0 | 4 | 2 | 1 | 0 | NA | Y |
| 13 | M | 73 | 0 | 3 | 1 | 1 | 0 | NA | Y |
| 17 | M | 58 | 0 | 4 | 2 | 0 | 0 | Y | Y |
| 20 | F | 75 | 1 | 2 | 0 | 0 | 2 | N | - |
| 25 | M | 65 | 0 | 3 | 0 | 0 | 0 | Y | - |
| 40 | M | 69 | 0 | 2 | 0 | 0 | 0 | N | - |
| 46 | M | 76 | 0 | 3 | 0 | 0 | 0 | Y | - |
| 48 | F | 80 | 0 | 3 | 0 | 1 | 0 | NA | Y |
| 78 | F | 54 | 0 | 4 | 1 | 1 | 0 | NA | Y |
| 97 | M | 79 | 0 | 3 | 0 | 0 | 0 | N | - |
| Ratio SW480/NCM460 | SSP | Identification | Peptides | Mascot Score | Mr (Da) | SwissProt Name |
|---|---|---|---|---|---|---|
| 0.15 | 9002 | Transgelin-2 | 2 | 48 | 22377 | TAGL2_HUMAN |
| 0.15 | 6204 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | 1 | 49 | 35793 | ECH1_HUMAN |
| 0.17 | 2804 | Transitional endoplasmic reticulum ATPase | 6 | 269 | 89958 | TERA_HUMAN |
| Src substrate protein p85 | 2 | 80 | 63462 | SRC8_HUMAN | ||
| 0.20 | 4001 | Protein S100-A4 | 2 | 125 | 11721 | S10A4_HUMAN |
| 0.24 | 1801 | Endoplasmin fragment | 9 | 405 | 92696 | ENPL_HUMAN |
| 0.26 | 4109 | Endoplasmic reticulum protein ERp29 | 1 | 75 | 28975 | ERP29_HUMAN |
| 0.27 | 2902 | Hypoxia up-regulated protein 1 | 11 | 401 | 111494 | HYOU1_HUMAN |
| 0.28 | 3601 | FK506-binding protein 4 | 6 | 366 | 52057 | FKBP4_HUMAN |
| 0.28 | 3705 | Heat shock cognate 71 kDa protein | 3 | 170 | 70854 | HSP7C_HUMAN |
| Lamin-B2 | 1 | 110 | 67647 | MMNB2_HUMAN | ||
| Stress-70 protein mitochondrial | 3 | 87 | 73635 | GRP75_HUMAN | ||
| 0.29 | 8608 | Adenylyl cyclase-associated protein 1 | 1 | 46 | 51823 | CAP1_HUMAN |
| 0.33 | 8701 | Heterogeneous nuclear ribonucleoprotein L | 3 | 70 | 60719 | HNRPL_HUMAN |
| 0.35 | 1501 | Tubulin beta | 3 | 229 | ca. 50000 | TBB2C_HUMAN ** |
| 0.38 | 1002 | Thioredoxin | 1 | 52 | 12015 | THIO_HUMAN |
| 0.39 | 9003 | Growth factor receptor-bound protein 2, fragment | 3 | 75 | 25190 | GRB2_HUMAN |
| 0.42 | 9207 | Voltage-dependent anion-selective channel protein 1 | 1 | 107 | 30722 | VDAC1_HUMAN |
| 0.42 | 1603 | Tubulin alpha | 4 | 195 | ca. 50000 | TBA1B_HUMAN ** |
| 0.43 | 5303 | 60S acidic ribosomal protein P0-like | 1 | 56 | 34343 | RLA0L_HUMAN |
| 0.45 | 8206 | Annexin A2 | 1 | 53 | 38580 | ANXA2_HUMAN |
| Putative annexin A2-like protein | 38635 | AXA2L_HUMAN | ||||
| 0.46 | 8204 | Annexin A2 | 2 | 120 | 38580 | ANXA2_HUMAN |
| Putative annexin A2-like protein | 38635 | AXA2L_HUMAN | ||||
| 0.46 | 6603 | T-complex protein 1 subunit beta | 6 | 418 | 57452 | TCPB_HUMAN |
| 0.46 | 6202 | Proteasome subunit alpha type-1 | 2 | 70 | 29537 | PSA1_HUMAN |
| 0.48 | 7403 | Elongation factor Tu, mitochondrial | 1 | 42 | 49510 | EFTU_HUMAN |
| 2.07 | 3206 | Tubulin alpha, fragment | 1 | 64 | ca. 50000 | TBA1B_HUMAN ** |
| 2.07 | 0401 | Putative heat chock protein HSP 90-beta-3, fragment | 1 | 42 | 68282 | H90B3_HUMAN |
| 2.11 | 3104 | Prohibitin | 4 | 131 | 29786 | PHB_HUMAN |
| 2.14 | 0301 | Nucleophosmin | 1 | 56 | 32555 | NPM_HUMAN |
| 2.16 | 0304 | Protein SET | 1 | 60 | 33469 | SET_HUMAN |
| 2.26 | 0607 | Endoplasmin, fragment | 4 | 170 | 92411 | ENPL_HUMAN |
| 2.76 | 0101 | Acidic leucine-rich nuclear phosphoprotein 32 family member A | 1 | 89 | 28568 | AN32A_HUMAN |
| 3.17 | 5003 | Vesicle-associated membrane protein-associated protein B/C, fragment | 4* | 156 | 27211 | VAPB_HUMAN |
| 3.28 | 3202 | Tubulin beta, fragment | 1 | 65 | ca. 50000 | TBB5_HUMAN ** |
| 6.08 | 3002 | Cellular retinoic acid-binding protein 2 | 7* | 401 | 15683 | RABP2_HUMAN |
| 6.65 | 4004 | Cofilin-1, fragment | 3 | 158 | 18507 | COF1_HUMAN |
| 8.58 | 5105 | Triosephosphate isomerase | 4 | 274 | 26698 | TPIS_HUMAN |
| 17.94 | 0008 | Nucleophosmin, fragment | 1 | 77 | 32555 | NPM_HUMAN |
| 18.45 | 3208 | ATP-dependent DNA helicase 2 subunit 2, fragment | 1 | 88 | 82652 | XRCC5_HUMAN |
| 99.68 | 0009 | Nucleophosmin, fragment | 1 | 48 | 32555 | NPM_HUMAN |
| 378.64 | 1007 | Retinol-binding protein I, cellular | 1 | 69 | 15840 | RET1_HUMAN |
| 3 * | 182 | |||||
| 4 * | 147 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludvigsen, M.; Thorlacius-Ussing, L.; Vorum, H.; Moyer, M.P.; Stender, M.T.; Thorlacius-Ussing, O.; Honoré, B. Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 3466. https://doi.org/10.3390/ijms21103466
Ludvigsen M, Thorlacius-Ussing L, Vorum H, Moyer MP, Stender MT, Thorlacius-Ussing O, Honoré B. Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer. International Journal of Molecular Sciences. 2020; 21(10):3466. https://doi.org/10.3390/ijms21103466
Chicago/Turabian StyleLudvigsen, Maja, Louise Thorlacius-Ussing, Henrik Vorum, Mary Pat Moyer, Mogens Tornby Stender, Ole Thorlacius-Ussing, and Bent Honoré. 2020. "Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer" International Journal of Molecular Sciences 21, no. 10: 3466. https://doi.org/10.3390/ijms21103466
APA StyleLudvigsen, M., Thorlacius-Ussing, L., Vorum, H., Moyer, M. P., Stender, M. T., Thorlacius-Ussing, O., & Honoré, B. (2020). Proteomic Characterization of Colorectal Cancer Cells versus Normal-Derived Colon Mucosa Cells: Approaching Identification of Novel Diagnostic Protein Biomarkers in Colorectal Cancer. International Journal of Molecular Sciences, 21(10), 3466. https://doi.org/10.3390/ijms21103466






