Modeling Direct and Indirect Action on Cell Survival After Photon Irradiation under Normoxia and Hypoxia
Abstract
1. Introduction
2. Results
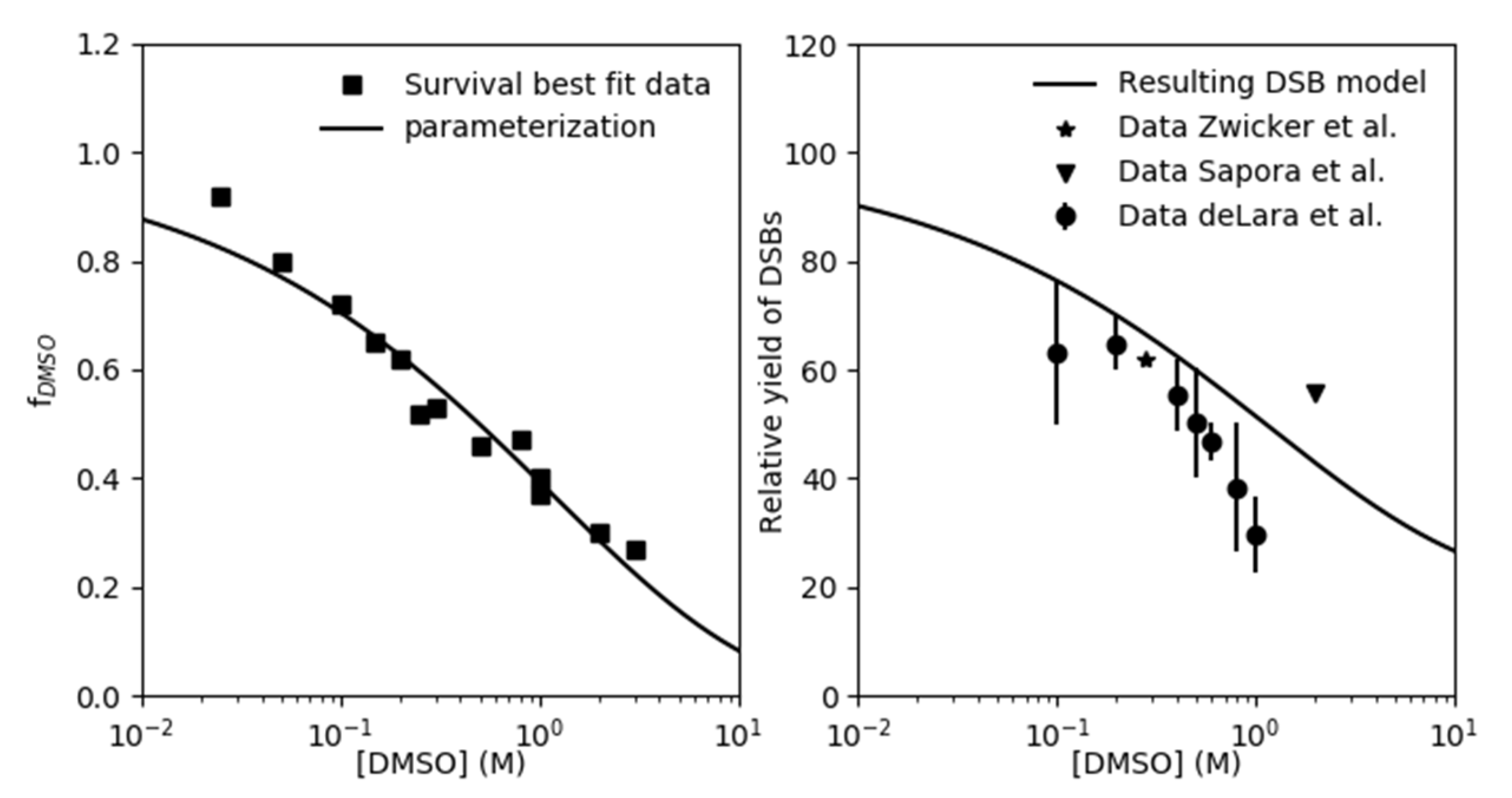
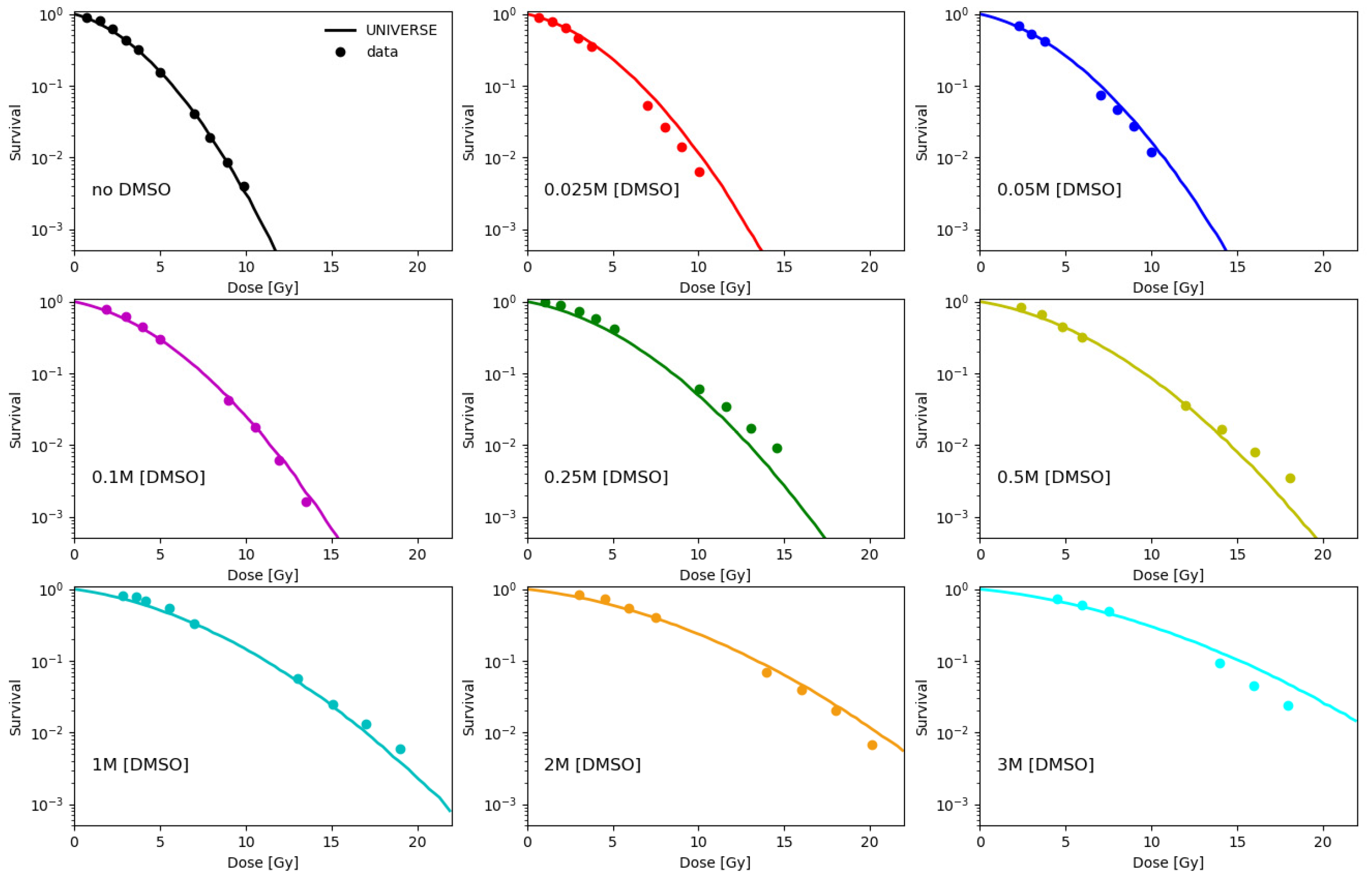
2.1. Parametrization of the Effect of DMSO on the Indirect Damage in Our Model
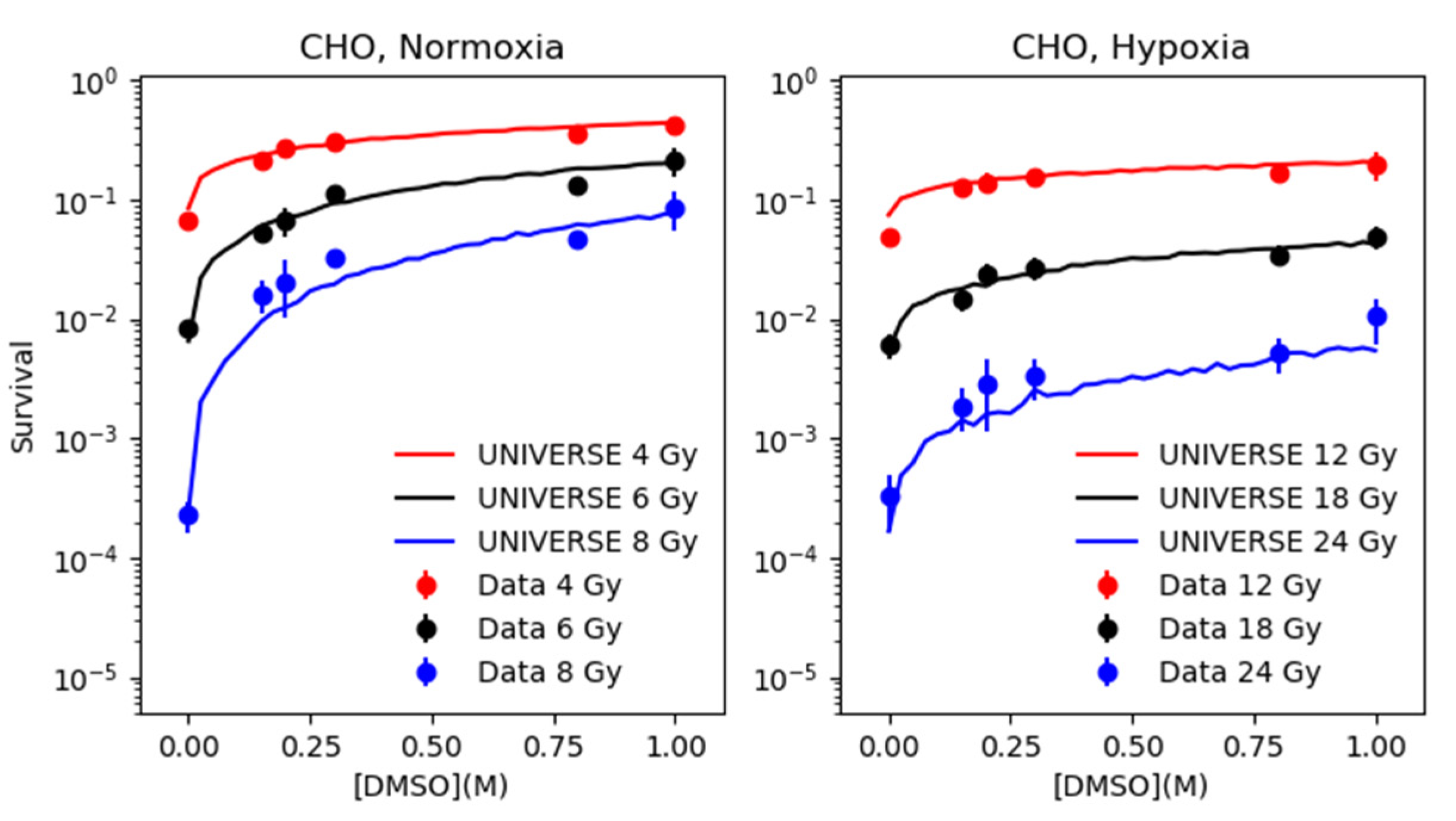
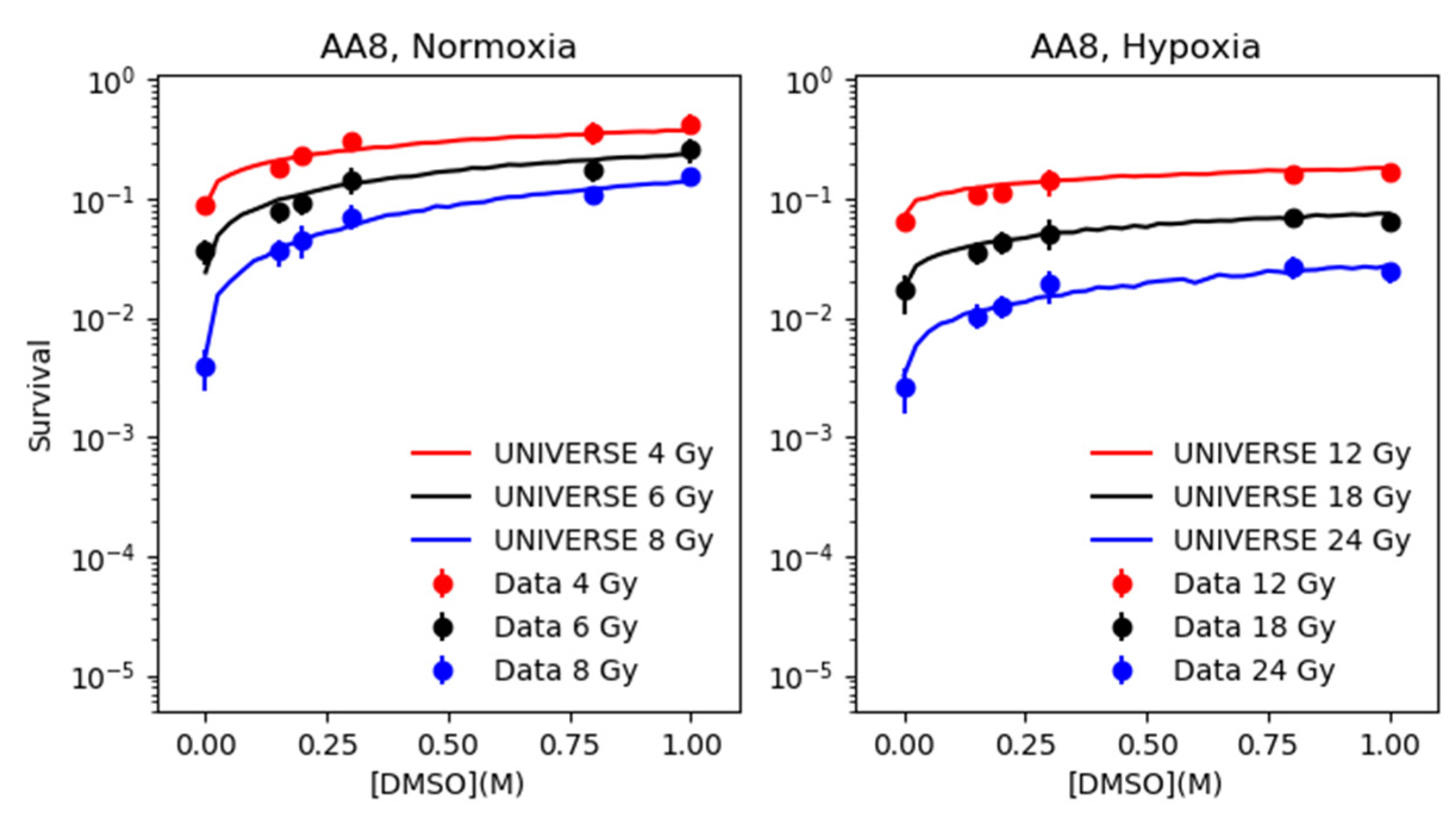
2.2. Modeling the Effect of DMSO under Normoxia and Hypoxia
3. Discussion
4. Materials and Methods
4.1. Experimental Data from Literature
4.2. Modeling Approach
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| LET | Linear energy transfer |
| DMSO | Dimethyl sulfoxide |
| UNIVERSE | UNIfied and VERSatile bio response Engine |
| DSB | (DNA) Double strand break |
| iDSB | Isolated DSB |
| cDSB | Complex DSB |
| OER | Oxygen enhancement ratio |
| HRF | Hypoxia reduction factor |
| CHO | Chinese hamster ovary |
References
- Delaney, G.; Jacob, S.; Featherstone, C.; Barton, M. The role of radiotherapy in cancer treatment: Estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer 2005, 104, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Radiobiological Modelling in Radiation Oncology; Dale, R.G., Jones, B., Eds.; British Institute of Radiology: London, UK, 2007; ISBN 978-0-905749-60-0. [Google Scholar]
- Liew, H.; Klein, C.; Zenke, F.T.; Abdollahi, A.; Debus, J.; Dokic, I.; Mairani, A. Modeling the Effect of Hypoxia and DNA Repair Inhibition on Cell Survival After Photon Irradiation. Int. J. Mol. Sci. 2019, 20, 6054. [Google Scholar] [CrossRef]
- Batey, M.A.; Zhao, Y.; Kyle, S.; Richardson, C.; Slade, A.; Martin, N.M.B.; Lau, A.; Newell, D.R.; Curtin, N.J. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol. Cancer 2013, 12, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, A.J.C.; Qiao, X.; Duursma, A.; Wijdeven, R.H.; Lieftink, C.; Hageman, F.; Morris, B.; Halonen, P.; Vens, C.; van den Brekel, M.W.M.; et al. Identification of a novel ATM inhibitor with cancer cell specific radiosensitization activity. Oncotarget 2017, 8, 73925–73937. [Google Scholar] [CrossRef] [PubMed]
- Durant, S.T.; Zheng, L.; Wang, Y.; Chen, K.; Zhang, L.; Zhang, T.; Yang, Z.; Riches, L.; Trinidad, A.G.; Fok, J.H.L.; et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci. Adv. 2018, 4, eaat1719. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.; Dokic, I.; Mairani, A.; Mein, S.; Brons, S.; Häring, P.; Haberer, T.; Jäkel, O.; Zimmermann, A.; Zenke, F.; et al. Overcoming hypoxia-induced tumor radioresistance in non-small cell lung cancer by targeting DNA-dependent protein kinase in combination with carbon ion irradiation. Radiat. Oncol. 2017, 12, 208. [Google Scholar] [CrossRef]
- Basic Clinical Radiobiology, 5th ed.; Joiner, M., van der Kogel, A., Eds.; CRC Press/Taylor & Francis Group: Boca Raton, FL, USA, 2018; ISBN 978-1-4441-7963-7. [Google Scholar]
- Tawk, B.; Schwager, C.; Deffaa, O.; Dyckhoff, G.; Warta, R.; Linge, A.; Krause, M.; Weichert, W.; Baumann, M.; Herold-Mende, C.; et al. Comparative analysis of transcriptomics based hypoxia signatures in head- and neck squamous cell carcinoma. Radiother. Oncol. 2016, 118, 350–358. [Google Scholar] [CrossRef]
- Rofstad, E.K.; Sundfør, K.; Lyng, H.; Tropé, C.G. Hypoxia-induced treatment failure in advanced squamous cell carcinoma of the uterine cervix is primarily due to hypoxia-induced radiation resistance rather than hypoxia-induced metastasis. Br. J. Cancer 2000, 83, 354–359. [Google Scholar] [CrossRef]
- Azzam, E.I.; Jay-Gerin, J.-P.; Pain, D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett. 2012, 327, 48–60. [Google Scholar] [CrossRef]
- Hirayama, R.; Ito, A.; Noguchi, M.; Matsumoto, Y.; Uzawa, A.; Kobashi, G.; Okayasu, R.; Furusawa, Y. OH radicals from the indirect actions of X-rays induce cell lethality and mediate the majority of the oxygen enhancement effect. Radiat. Res. 2013, 180, 514–523. [Google Scholar] [CrossRef]
- Johnke, R.M.; Sattler, J.A.; Allison, R.R. Radioprotective agents for radiation therapy: Future trends. Future Oncol. 2014, 10, 2345–2357. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Cramer, H.M.; Inch, W.R.; Lampe, H.B. Radioprotective effect of free radical scavenging enzymes. J. Otolaryngol. 1990, 19, 299–306. [Google Scholar] [PubMed]
- Smith, T.A.; Kirkpatrick, D.R.; Smith, S.; Smith, T.K.; Pearson, T.; Kailasam, A.; Herrmann, K.Z.; Schubert, J.; Agrawal, D.K. Radioprotective agents to prevent cellular damage due to ionizing radiation. J. Transl. Med. 2017, 15, 232. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.D.; Doern, S.D.; Reuvers, A.P.; Gillespie, C.J.; Chatterjee, A.; Blakely, E.A.; Smith, K.C.; Tobias, C.A. Radioprotection by DMSO of mammalian cells exposed to X-rays and to heavy charged-particle beams. Radiat. Env. Biophys. 1979, 16, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Bishayee, A.; Rao, D.V.; Bouchet, L.G.; Bolch, W.E.; Howell, R.W. Protection by DMSO against Cell Death Caused by Intracellularly Localized Iodine-125, Iodine-131 and Polonium-210. Radiat. Res. 2000, 153, 416–427. [Google Scholar] [CrossRef][Green Version]
- Kim, S.E.; Moos, W.S. Radiation protection by topical DMSO application. Health Phys. 1967, 13, 601–606. [Google Scholar] [CrossRef]
- Willson, R.L. The reaction of oxygen with radiation-induced free radicals in DNA and related compounds. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1970, 17, 349–358. [Google Scholar] [CrossRef]
- Alexander, P. Division of Biophysics: On the Mode of Action of Some Treatments That Influence the Radiation Sensitivity of Cells. Trans. New York Acad. Sci. 1962, 24, 966–978. [Google Scholar] [CrossRef]
- Sapora, O.; Barone, F.; Belli, M.; Maggi, A.; Quintiliani, M.; Tabocchini, M.A. Relationships Between Cell Killing, Mutation Induction and DNA Damage in X-irradiated V79 Cells: The Influence of Oxygen and DMSO. Int. J. Radiat. Biol. 1991, 60, 467–482. [Google Scholar] [CrossRef]
- Frankenberg, D.; Frankenberg-Schwager, M.; Harbich, R. Mechanisms of Oxygen Radiosensitization in Irradiated Yeast. Int. J. Radiat. Biol. 1993, 64, 511–521. [Google Scholar] [CrossRef]
- deLara, C.M.; Jenner, T.J.; Townsend, K.M.; Marsden, S.J.; O’Neill, P. The effect of dimethyl sulfoxide on the induction of DNA double-strand breaks in V79-4 mammalian cells by alpha particles. Radiat. Res. 1995, 144, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Zwicker, F.; Hauswald, H.; Debus, J.; Huber, P.E.; Weber, K.-J. Impact of dimethyl sulfoxide on irradiation-related DNA double-strand-break induction, -repair and cell survival. Radiat. Env. Biophys. 2019, 58, 417–424. [Google Scholar] [CrossRef]
- Ewing, D.; Walton, H.L. Radiation protection of in vitro mammalian cells: Effects of hydroxyl radical scavengers on the slopes and shoulders of survival curves. Radiat. Res. 1991, 126, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Kashino, G.; Liu, Y.; Suzuki, M.; Masunaga, S.; Kinashi, Y.; Ono, K.; Tano, K.; Watanabe, M. An alternative mechanism for radioprotection by dimethyl sulfoxide; possible facilitation of DNA double-strand break repair. J. Radiat. Res. 2010, 51, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Mairani, A.; Böhlen, T.T.; Dokic, I.; Cabal, G.; Brons, S.; Haberer, T. Modelling of cell killing due to sparsely ionizing radiation in normoxic and hypoxic conditions and an extension to high LET radiation. Int. J. Radiat. Biol. 2013, 89, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Frankenberg-Schwager, M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother. Oncol. 1989, 14, 307–320. [Google Scholar] [CrossRef]
- Prise, K.M.; Ahnström, G.; Belli, M.; Carlsson, J.; Frankenberg, D.; Kiefer, J.; Löbrich, M.; Michael, B.D.; Nygren, J.; Simone, G.; et al. A review of dsb induction data for varying quality radiations. Int. J. Radiat. Biol. 1998, 74, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Prise, K.M.; Pinto, M.; Newman, H.C.; Michael, B.D. A review of studies of ionizing radiation-induced double-strand break clustering. Radiat. Res. 2001, 156, 572–576. [Google Scholar] [CrossRef]
- Asaithamby, A.; Chen, D.J. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009, 37, 3912–3923. [Google Scholar] [CrossRef]
- Elsässer, T.; Weyrather, W.K.; Friedrich, T.; Durante, M.; Iancu, G.; Krämer, M.; Kragl, G.; Brons, S.; Winter, M.; Weber, K.-J.; et al. Quantification of the relative biological effectiveness for ion beam radiotherapy: Direct experimental comparison of proton and carbon ion beams and a novel approach for treatment planning. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 1177–1183. [Google Scholar] [CrossRef]
- Friedrich, T.; Durante, M.; Scholz, M. Modeling cell survival after photon irradiation based on double-strand break clustering in megabase pair chromatin loops. Radiat. Res. 2012, 178, 385–394. [Google Scholar] [CrossRef]
- Friedrich, T.; Scholz, U.; Elsässer, T.; Durante, M.; Scholz, M. Calculation of the biological effects of ion beams based on the microscopic spatial damage distribution pattern. Int. J. Radiat. Biol. 2012, 88, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Yokota, H.; van den Engh, G.; Hearst, J.E.; Sachs, R.K.; Trask, B.J. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J. Cell Biol. 1995, 130, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Sachs, R.K.; van den Engh, G.; Trask, B.; Yokota, H.; Hearst, J.E. A random-walk/giant-loop model for interphase chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 2710–2714. [Google Scholar] [CrossRef] [PubMed]
- Solovjeva, L.; Svetlova, M.; Stein, G.; Chagin, V.; Rozanov, Y.; Zannis-Hadjopoulos, M.; Price, G.; Tomilin, N. Conformation of replicated segments of chromosome fibres in human S-phase nucleus. Chromosome Res. 1998, 6, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Carlson, D.J.; Stewart, R.D.; Semenenko, V.A. Effects of oxygen on intrinsic radiation sensitivity: A test of the relationship between aerobic and hypoxic linear-quadratic (LQ) model parameters. Med. Phys. 2006, 33, 3105–3115. [Google Scholar] [CrossRef]
- Alper, T.; Howard-Flanders, P. Role of Oxygen in Modifying the Radiosensitivity of E. Coli B. Nature 1956, 178, 978–979. [Google Scholar] [CrossRef]
| Cell Line | Reference | |||
|---|---|---|---|---|
| CHO | 5.56 × 10−3 ± 1.12 × 10−3 | 7.65 × 10−1 ± 0.44 × 10−1 | 2.90 | Hirayama et al. 2013 [12] |
| AA8 | 14.00 × 10−3 ± 1.48 × 10−3 | 9.16 × 10−1 ± 0.89 × 10−1 | 2.85 | Hirayama et al. 2013 [12] |
| V79 | 4.79 × 10−3 ± 0.52 × 10−3 | 3.17 × 10−1 ± 0.13 × 10−1 | – | Chapman et al. 1979 [16] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liew, H.; Mein, S.; Debus, J.; Dokic, I.; Mairani, A. Modeling Direct and Indirect Action on Cell Survival After Photon Irradiation under Normoxia and Hypoxia. Int. J. Mol. Sci. 2020, 21, 3471. https://doi.org/10.3390/ijms21103471
Liew H, Mein S, Debus J, Dokic I, Mairani A. Modeling Direct and Indirect Action on Cell Survival After Photon Irradiation under Normoxia and Hypoxia. International Journal of Molecular Sciences. 2020; 21(10):3471. https://doi.org/10.3390/ijms21103471
Chicago/Turabian StyleLiew, Hans, Stewart Mein, Jürgen Debus, Ivana Dokic, and Andrea Mairani. 2020. "Modeling Direct and Indirect Action on Cell Survival After Photon Irradiation under Normoxia and Hypoxia" International Journal of Molecular Sciences 21, no. 10: 3471. https://doi.org/10.3390/ijms21103471
APA StyleLiew, H., Mein, S., Debus, J., Dokic, I., & Mairani, A. (2020). Modeling Direct and Indirect Action on Cell Survival After Photon Irradiation under Normoxia and Hypoxia. International Journal of Molecular Sciences, 21(10), 3471. https://doi.org/10.3390/ijms21103471




