A Novel Protocol for Detection of Senescence and Calcification Markers by Fluorescence Microscopy
Abstract
1. Introduction
2. Results and Discussion
2.1. Detection of SA-β-Gal
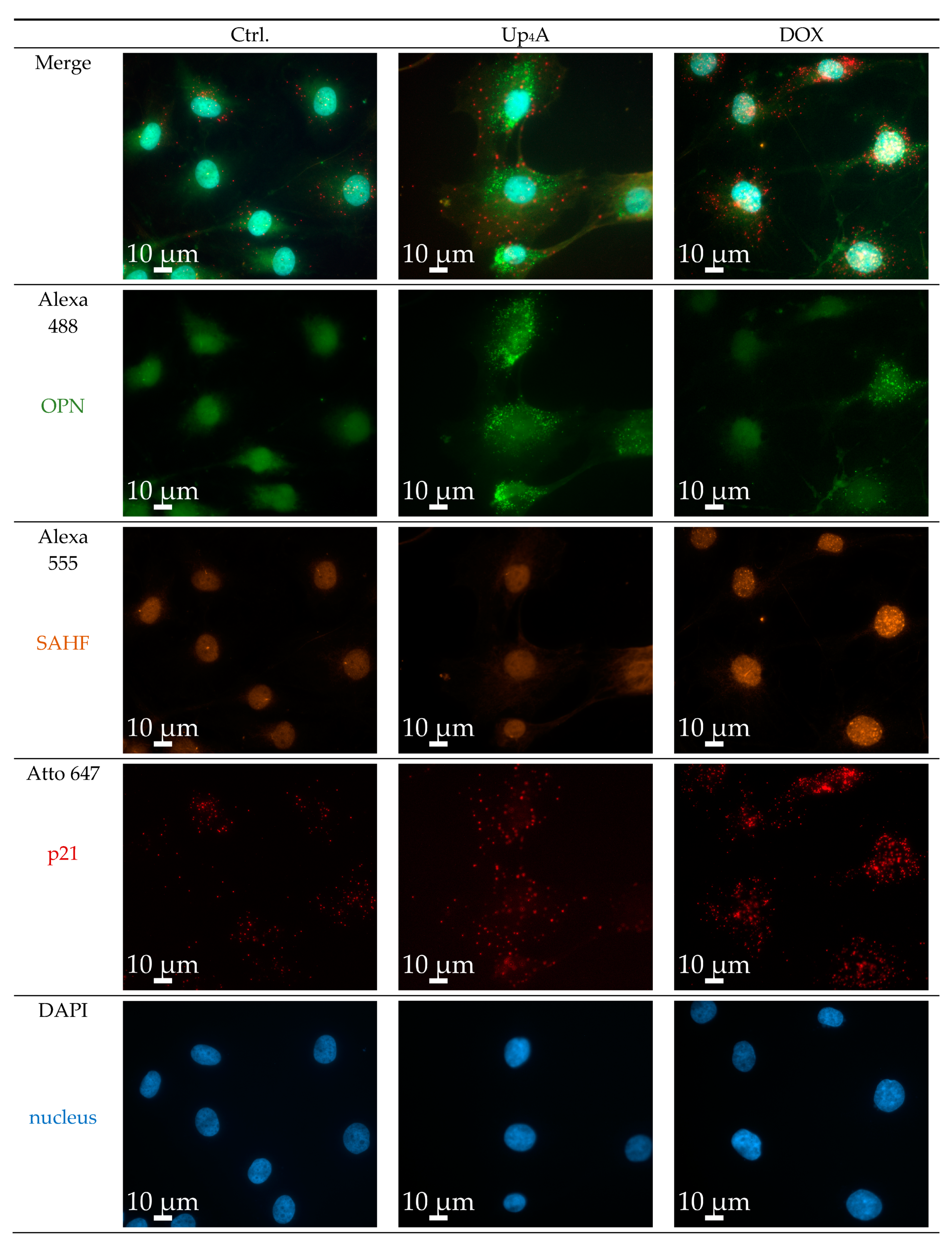
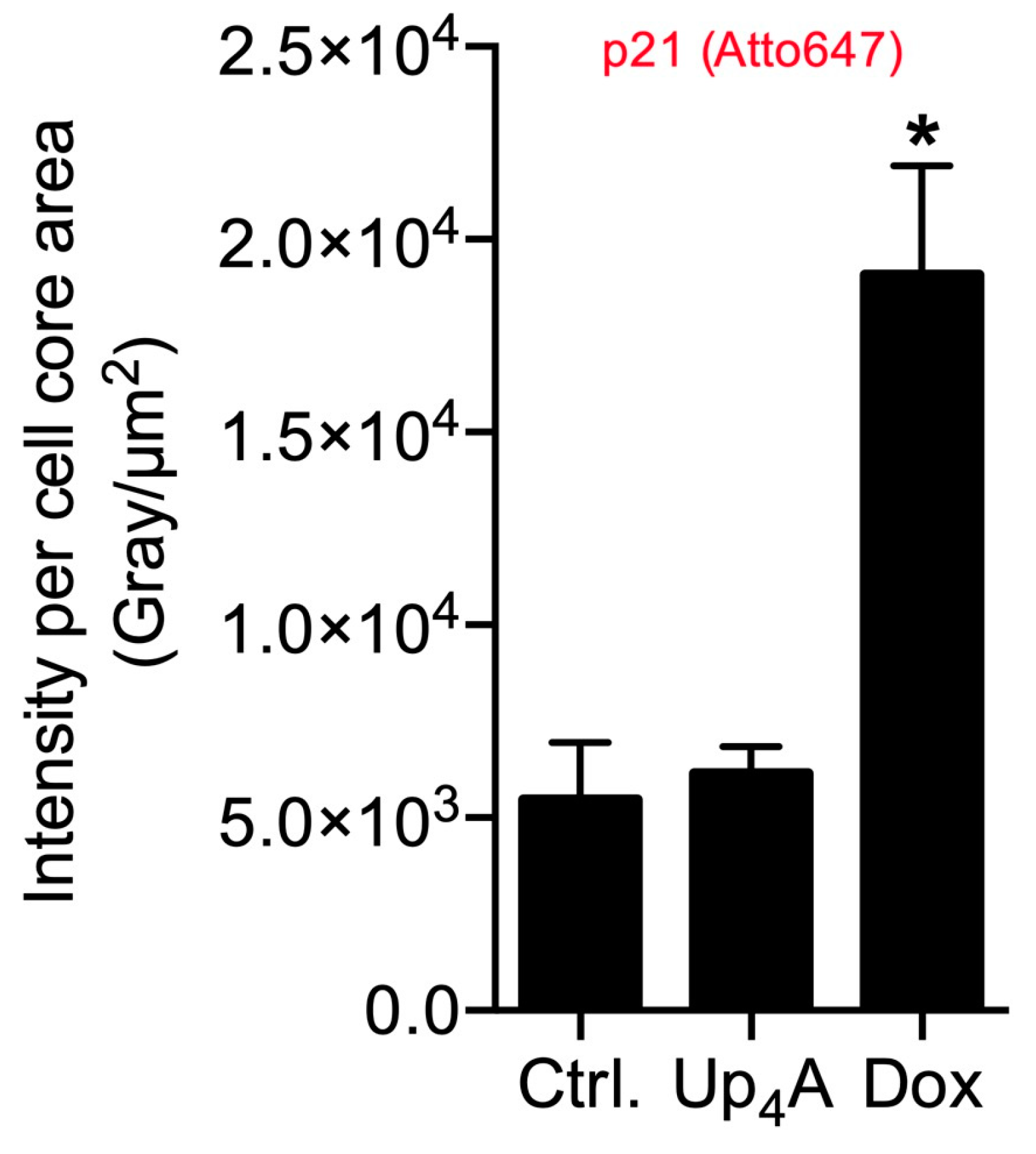
2.2. OPN and p21 mRNA Detection and Detection of SAHF
2.3. Limitations
3. Materials and Methods
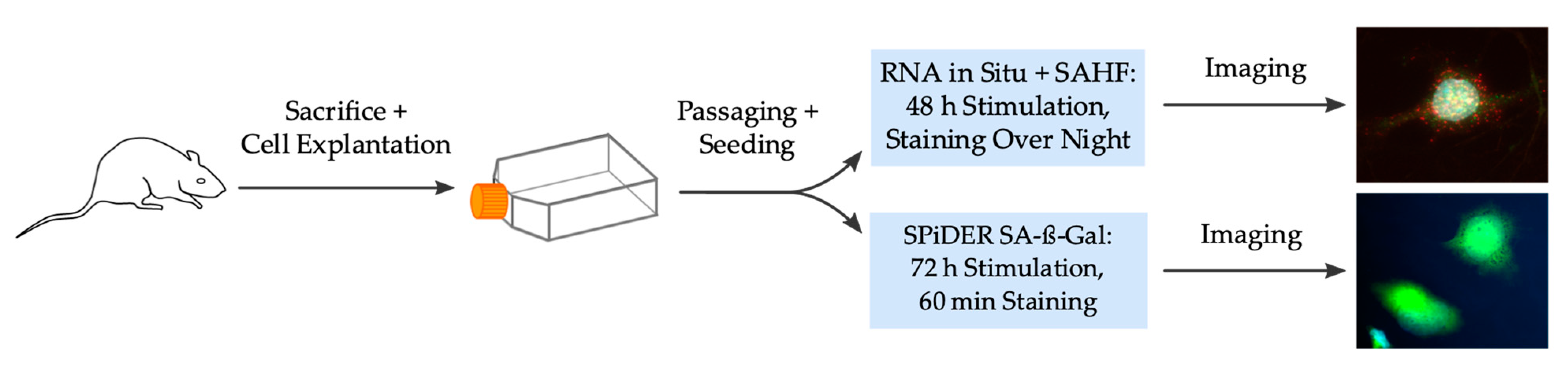
3.1. Cell Isolation and Culturing
3.2. Experimental Setting for Detection of SA-β-Galactosidase
3.2.1. Preparations
- Heat incubator to 37 °C (no humidity and carbon dioxide control).
- Warm 4% formalin and PBS to 37 °C.
- Prepare McIlvaine buffer: Mix 7.4 mL 0.1 mol/L citric acid solution and 12.6 mL 0.2 mol/L sodium phosphate solution and set pH to 6.0.
- Solve 20 µg SPiDER-SA-β-Gal (Gerbu Biotechnologie) in 35 µL dimethylsulfoxide (DMSO). Store aliquots at −20°C.
- Dilute McIlvaine buffer 1:5 in ultrapure water and warm dilution to 37 °C. Dilute SPiDER-SA-ß-Gal 1:500 in McIlvaine buffer in order to obtain the working solution. Protect working solution from light.
- Prepare Hoechst working solution by dissolving Hoechst 33342 (Thermo Fisher) in the appropriate amount of water to obtain a stock concentration of 10 mg/mL. The stock concentration can be aliquoted and stored at −20°C. To obtain the working solution, dilute Hoechst 1:2000 in PBS. Protect working solution from light.
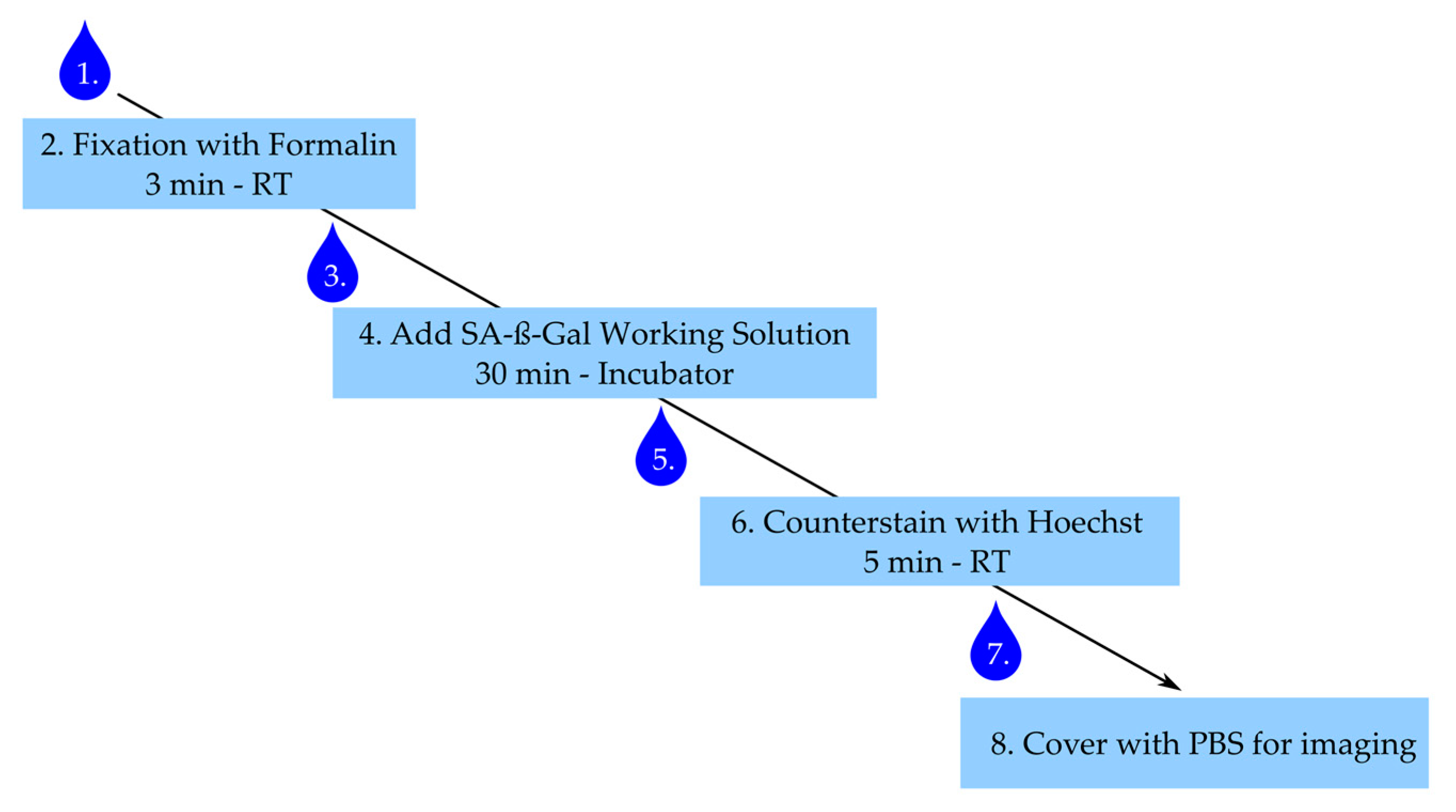
3.2.2. Staining Procedure
- After stimulation, aspirate medium and wash cells once with PBS.
- Add 300 µL of 4% buffered formalin to each well and fix cells for 3 min at room temperature, ensure the exact fixation time.
- Aspirate formalin and wash three times with warm PBS.
- Add 300 µL of working solution per well and incubate for 30 min in the incubator under light protection. Caution: Ensure the right fixation times and pH conditions—this is critical for the β-Gal staining.
- Aspirate working solution and wash cells twice with PBS.
- Counterstain with Hoechst working solution for 5 min under light protection.
- Aspirate solution and wash once with PBS.
- Add 300 µL of PBS and image within 24 h.
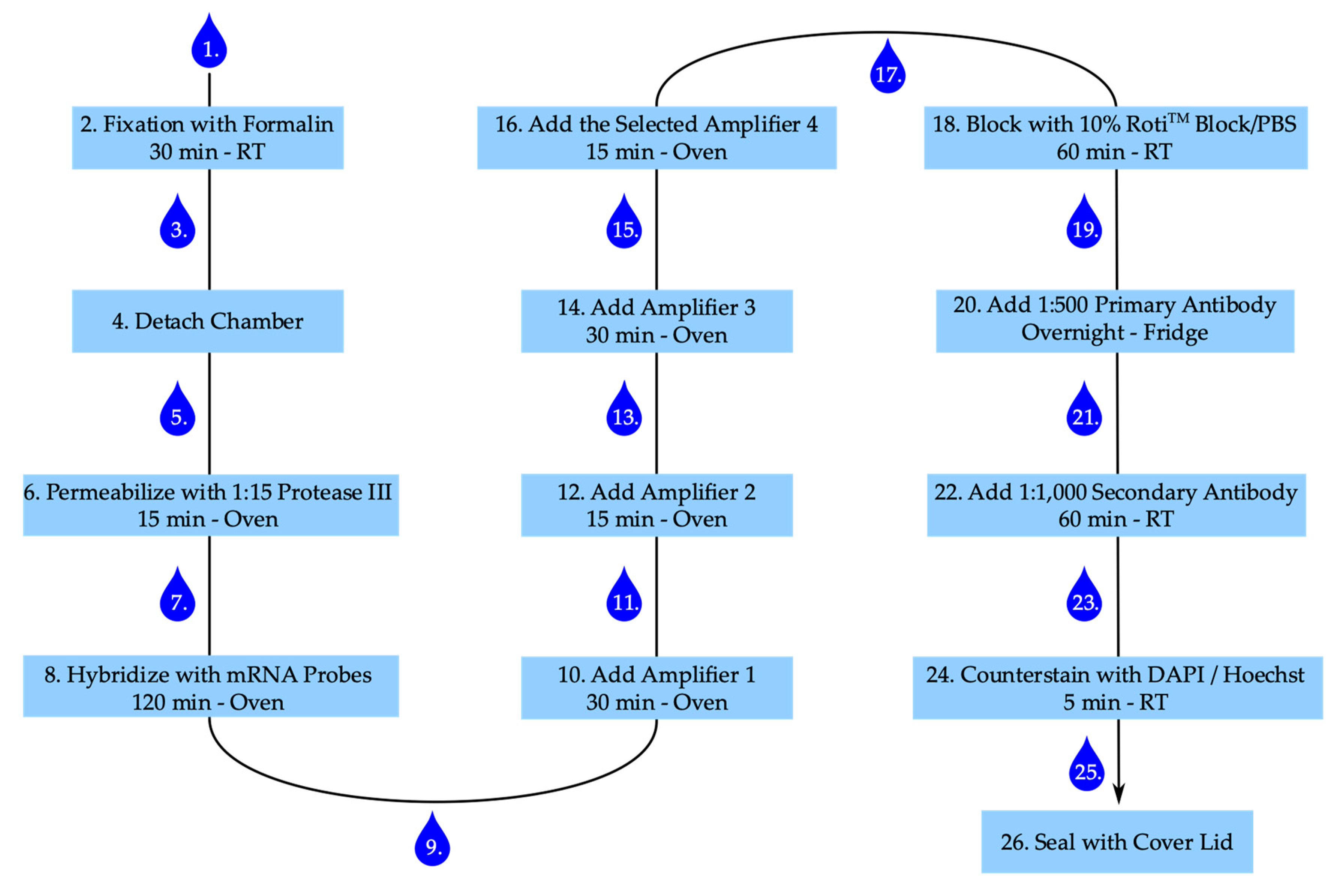
3.3. Experimental Setting for Detection of OPN, p21 and SAHF
3.3.1. Preparations
- Add purified water to the humidity control tray and heat oven to 40 °C.
- Thaw ProLong™ diamond antifade medium (Thermo Scientific) at room temperature.
- Prepare wash buffer (ACD Bio) according to manufacturer’s instructions.
- Heat mRNA probes (ACD Bio) gently at 40 °C for 10 min in a water bath, centrifuge probes and mix according to manufacturer’s instructions.
- Dilute Protease III (ACD Bio) 1:15 with PBS.
- Prepare washing dishes for PBS and wash buffer.
- Prepare 10% Roti™ImmunoBlock (Carl Roth) by diluting in PBS.
- Prepare 1:500 dilution of primary antibody anti-histone H2A.X (phosphoS139) antibody [EP854(2)Y] (abcam) in 1% Roti™ImmunoBlock/PBS.
- Prepare 1:1,000 dilution of secondary goat anti-Rabbit IgG (H+L) highly cross-adsorbed antibody, Alexa Fluor 555 (Invitrogen) in 1% Roti™ImmunoBlock/PBS.
- If applicable: Prepare Hoechst stain, as explained above.
3.3.2. Staining Procedure
- After stimulation aspirate medium and wash cells once with PBS.
- Add 300 µL of 4% buffered formalin to each well and fix cells for 30 min at room temperature.
- Aspirate formalin and wash twice with PBS.
- Carefully detach the chamber from the slide according to the manufacturer’s instruction and place the slide in a PBS filled washing dish. Caution: The glue is strong. Make sure to remove the glue of the chamber properly, otherwise the slide–coverslip combination becomes too thick.
- Remove slide from the washing dish and thoroughly apply a barrier around each well with the ImmEdge™ hydrophobic barrier pen (ACD Bio) and place slide again in PBS.
- Remove the slide from PBS, remove attaching PBS by gently inverting the slide and add 50 µL of diluted Protease III to each well. Place slide in the humidity control tray, close humidity control tray, and incubate in the oven for 15 min.
- Remove slides from tray; remove protease from slide by inverting the slide and place slide in fresh PBS. Caution: The movement should be gently, but still removing the majority of liquid.
- Remove slide from PBS, remove PBS by gently inverting the slide, and add 50 µL of diluted target probes or one drop of positive or negative control to the according wells, place slides in the humidity control tray, and incubate in the oven for 120 min.
- Take slides out of the tray, inverse, and wash twice for 2 min each in wash buffer.
- Remove attached liquid by gentle inversion and add one drop of amplifier 1-fluid (Amp 1-FL) to each well. Put slides in the humidity control tray in the oven for 30 min.
- Take slides out of the tray, inverse, and wash twice for 2 min each in wash buffer.
- Remove attached liquid by gentle inversion and add 1 drop of Amp 2-FL to each well, place in the humidity control tray in the oven for 15 min.
- After 15 min take slides out of the tray, inverse and wash twice for 2 min each in wash buffer.
- Remove attached liquid by gentle inversion and add 1 drop of Amp 3-FL to each well, place in the humidity control tray in the oven for 30 min.
- Take slides out of the tray, inverse and wash twice for 2 min each in wash buffer.
- Remove attached liquid by gentle inversion and add 1 drop of the selected Amp 4-FL to each well, place in the humidity control tray in the oven for 15 min. Caution: For the multiplexing protocol we used Amp 4-FL A.
- Take slides out of the tray, inverse and wash twice for 2 min each in wash buffer.
- Add 100 µL of 10% Roti™ImmunoBlock/PBS to each well and block for 1 h in the closed humidity control tray at room temperature.
- Remove attached liquid by gentle inversion and wash once with PBS.
- Add 50 µL 1:500 dilution of primary anti-histone H2A.X (phosphoS139) antibody [EP854(2)Y] (Abcam) in 1% Roti™ImmunoBlock/PBS per well and incubate in the closed humidity control tray in the fridge overnight.
- Remove attached liquid by gentle inversion and wash twice with PBS.
- Add 50 µL 1:1,000 dilution of secondary goat anti-rabbit IgG (H+L) Highly Cross-Adsorbed antibody, Alexa Fluor 555 (Invitrogen) in 1% Roti™ImmunoBlock/PBS per well and incubate in the closed humidity control tray at room temperature for 60 min.
- Remove attached liquid by gentle inversion and wash twice with PBS.
- Add 1 drop of 4′,6-diamidino-2-phenylindole (DAPI) per well and incubate for 1 min at room temperature (alternatively add 50 µL 1:2,000 Hoechst working solution and incubate for 5 min at room temperature).
- Remove attached liquid by gentle inversion and wash once with PBS.
- Add two drops of ProLong™ diamond antifade medium (Thermo Scientific) to each slide and gently apply the lid, make sure to gently remove all bubbles, and harden overnight in the fridge. Caution: The medium is highly viscose and can easily dry out. The cover slid is still moveable, even after drying. Be careful when cleaning the slide for imaging. Sealing the slide-lid combination with nail varnish can help in preventing the drying out and preserving the slides for later imaging.
3.4. Imaging
3.5. Quantification of Fluorescence Intensity
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Amp-FL | Amplifier fluid |
| Ctrl | Control |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMSO | Dimethylsulfoxide |
| DNA | Deoxyribonucleic acid |
| DMEM | Dulbeccos-modified Eagle medium |
| Dox | Doxorubicin |
| FCS | Fetal calf serum |
| mRNA | Messenger Ribonucleic acid |
| OPN | Osteopontin |
| PCR | Polymerase chain reaction |
| PBS | Phosphate-buffered saline |
| RT | Room temperature |
| rVSMC | Rat vascular smooth muscle cell |
| SA-β-Gal | Senescence-associated β-galactosidase |
| SAHF | Senescence-associated heterochromatin foci |
| SEM | Standard error of mean |
| Up4A | Uridine adenosine tetraphosphate |
| VSMC | Vascular smooth muscle cell |
| X-Gal (BCIG) | 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside |
References
- Jaminon, A.; Reesink, K.; Kroon, A.; Schurgers, L. The Role of Vascular Smooth Muscle Cells in Arterial Remodeling: Focus on Calcification-Related Processes. Int. J. Mol. Sci. 2019, 20, 5694. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, P.M.; Lurbe, E.; Laurent, S. The early life origins of vascular ageing and cardiovascular risk: The EVA syndrome. J. Hypertens 2008, 26, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Voelkl, J.; Lang, F.; Eckardt, K.-U.; Amann, K.; Kuro-O, M.; Pasch, A.; Pieske, B.; Alesutan, I. Signaling pathways involved in vascular smooth muscle cell calcification during hyperphosphatemia. Cell Mol. Life Sci. 2019, 76, 2077–2091. [Google Scholar] [CrossRef] [PubMed]
- Henaut, L.; Mary, A.; Chillon, J.M.; Kamel, S.; Massy, Z.A. The Impact of Uremic Toxins on Vascular Smooth Muscle Cell Function. Toxins 2018, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Tolle, M.; Reshetnik, A.; Schuchardt, M.; Hohne, M.; van der Giet, M. Arteriosclerosis and vascular calcification: Causes, clinical assessment and therapy. Eur. J. Clin. Investig. 2015, 45, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Sanchis, P.; Ho, C.Y.; Liu, Y.; Beltran, L.E.; Ahmad, S.; Jacob, A.P.; Furmanik, M.; Laycock, J.; Long, D.A.; Shroff, R.; et al. Arterial “inflammaging” drives vascular calcification in children on dialysis. Kidney Int. 2019, 95, 958–972. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.; Babic, M.; Tolle, M.; van der Giet, M.; Schuchardt, M. Research Models for Studying Vascular Calcification. Int. J. Mol. Sci. 2020, 21, 2204. [Google Scholar] [CrossRef] [PubMed]
- Burton, D.G.; Matsubara, H.; Ikeda, K. Pathophysiology of vascular calcification: Pivotal role of cellular senescence in vascular smooth muscle cells. Exp. Gerontol. 2010, 45, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Muteliefu, G.; Shimizu, H.; Enomoto, A.; Nishijima, F.; Takahashi, M.; Niwa, T. Indoxyl sulfate promotes vascular smooth muscle cell senescence with upregulation of p53, p21, and prelamin A through oxidative stress. Am. J. Physiol. Cell Physiol. 2012, 303, C126–C134. [Google Scholar] [CrossRef] [PubMed]
- Tuttle, C.S.L.; Waaijer, M.E.C.; Slee-Valentijn, M.S.; Stijnen, T.; Westendorp, R.; Maier, A.B. Cellular senescence and chronological age in various human tissues: A systematic review and meta-analysis. Aging. Cell 2020, 19, e13083. [Google Scholar] [CrossRef] [PubMed]
- Carnero, A. Markers of cellular senescence. Methods Mol. Biol. (Cliftonn. J.) 2013, 965, 63–81. [Google Scholar]
- Noren Hooten, N.; Evans, M.K. Techniques to Induce and Quantify Cellular Senescence. J. Vis. Exp. 2017, 123, 55533. [Google Scholar] [CrossRef] [PubMed]
- Giachelli, C.M. Ectopic calcification: Gathering hard facts about soft tissue mineralization. Am. J. Pathol. 1999, 154, 671–675. [Google Scholar] [CrossRef]
- Jensen, E. Technical review: In situ hybridization. Anat. Rec. (Hoboken) 2014, 297, 1349–1353. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.T.; Ma, X.J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Cappetta, D.; De Angelis, A.; Sapio, L.; Prezioso, L.; Illiano, M.; Quaini, F.; Rossi, F.; Berrino, L.; Naviglio, S.; Urbanek, K. Oxidative Stress and Cellular Response to Doxorubicin: A Common Factor in the Complex Milieu of Anthracycline Cardiotoxicity. Oxid. Med. Cell Longev. 2017, 2017, 1521020. [Google Scholar] [CrossRef] [PubMed]
- Schuchardt, M.; Tolle, M.; Prufer, J.; Prufer, N.; Huang, T.; Jankowski, V.; Jankowski, J.; Zidek, W.; van der Giet, M. Uridine adenosine tetraphosphate activation of the purinergic receptor P2Y enhances in vitro vascular calcification. Kidney Int. 2012, 81, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.-C.; Hu, M.-L. The limitations and validities of senescence associated-β-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp. Gerontol. 2005, 40, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.M.S.; Burch, R.L. The Principles of Humane Experimental Technique; Methuen & Co Ltd.: London, UK, 1959. [Google Scholar]


| Dye | SPiDER SA-β-Gal | Hoechst 33342 | Alexa 488 | Alexa 555 | Atto 647 | DAPI |
|---|---|---|---|---|---|---|
| Beam Splitter | 532 | 395 | 495 | 570 | 660 | 395 |
| Filter Ex. Wavelength | 500–530 | 335–383 | 450–490 | 538–562 | 625–665 | 335–383 |
| Filter Em. Wavelength | 545–605 | 420–470 | 500–550 | 570–640 | 665–715 | 420–470 |
| Ex. Wavelength of Dye | 528 | 348 | 493 | 553 | 644 | 348 |
| Em. Wavelength of Dye | 547 | 455 | 517 | 568 | 670 | 455 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrmann, J.; Babic, M.; Tölle, M.; Eckardt, K.-U.; van der Giet, M.; Schuchardt, M. A Novel Protocol for Detection of Senescence and Calcification Markers by Fluorescence Microscopy. Int. J. Mol. Sci. 2020, 21, 3475. https://doi.org/10.3390/ijms21103475
Herrmann J, Babic M, Tölle M, Eckardt K-U, van der Giet M, Schuchardt M. A Novel Protocol for Detection of Senescence and Calcification Markers by Fluorescence Microscopy. International Journal of Molecular Sciences. 2020; 21(10):3475. https://doi.org/10.3390/ijms21103475
Chicago/Turabian StyleHerrmann, Jaqueline, Milen Babic, Markus Tölle, Kai-Uwe Eckardt, Markus van der Giet, and Mirjam Schuchardt. 2020. "A Novel Protocol for Detection of Senescence and Calcification Markers by Fluorescence Microscopy" International Journal of Molecular Sciences 21, no. 10: 3475. https://doi.org/10.3390/ijms21103475
APA StyleHerrmann, J., Babic, M., Tölle, M., Eckardt, K.-U., van der Giet, M., & Schuchardt, M. (2020). A Novel Protocol for Detection of Senescence and Calcification Markers by Fluorescence Microscopy. International Journal of Molecular Sciences, 21(10), 3475. https://doi.org/10.3390/ijms21103475





