Physical Activity Dynamically Regulates the Hippocampal Proteome along the Dorso-Ventral Axis
Abstract
:1. Introduction
2. Results
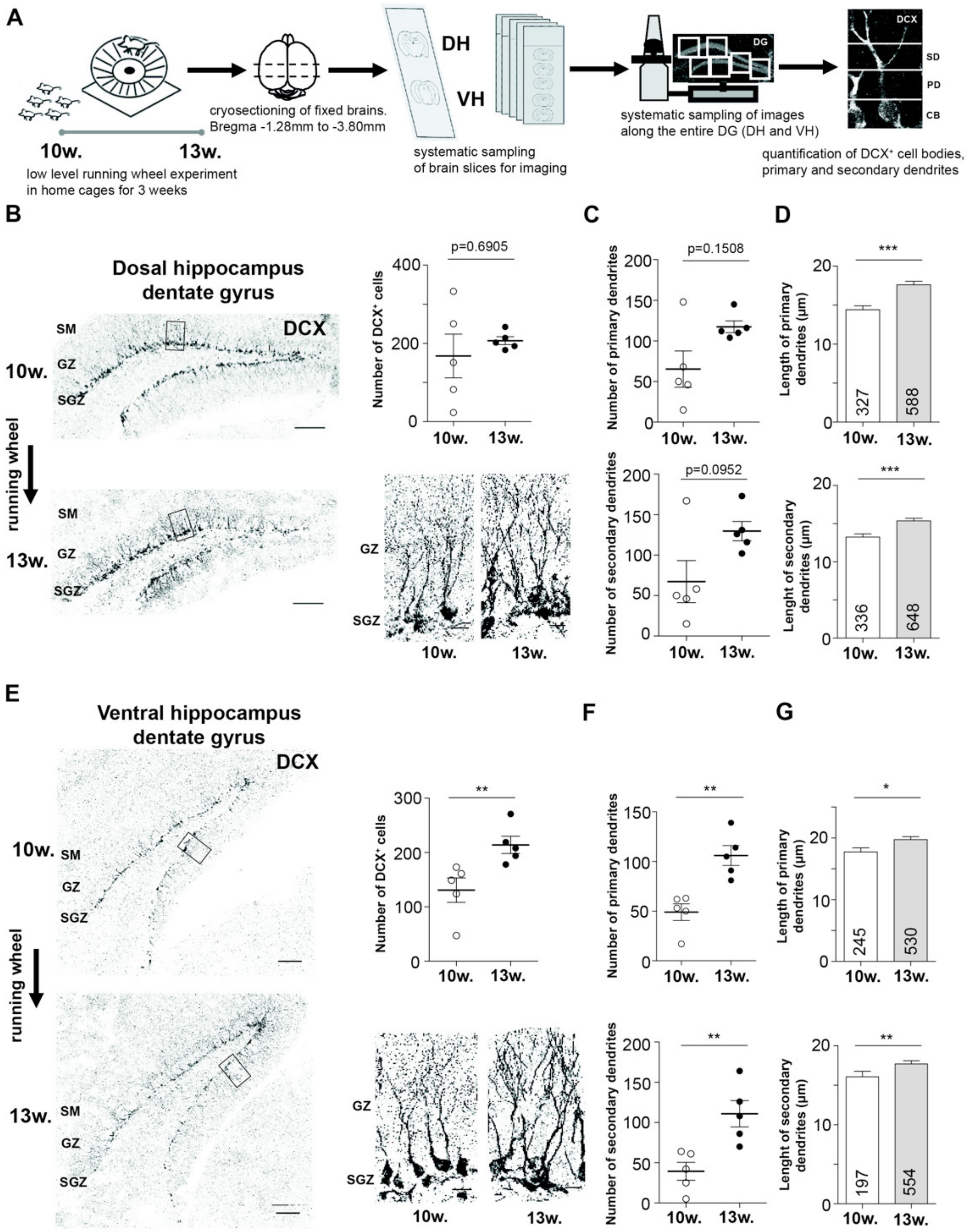
2.1. EPA Differently Impacts Adult Neurogenesis in Dorsal and Ventral Hippocampus
2.2. Label Free Quantitative Mass Spectrometry Detects Proteomic Alterations during Neurogenesis
2.3. Dorsal and Ventral Hippocampal Subregions Respond Differently to EPA
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Immunohistochemistry
4.2.1. Tissue Preparation
4.2.2. Tissue Selection
4.2.3. Imaging
4.2.4. Quantification
4.3. Proteomics
4.3.1. Tissue Preparation and Microdissection
4.3.2. Mass Spectrometry Analysis
4.3.3. Data Analysis
4.4. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CA | Cornu Ammonis |
| DCX | Doublecortin |
| DG | Dentate gyrus |
| EPA | Enhanced physical activity |
References
- Fanselow, M.S.; Dong, H. Are the Dorsal and Ventral Hippocampus Functionally Distinct Structures ? Neuron 2010, 65, 7–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moser, M.B.; Moser, E.I.; Forrest, E.; Andersen, P.; Morris, R.G.M. Spatial learning with a minislab in the dorsal hippocampus. Proc. Natl. Acad. Sci. USA 1995, 92, 9697–9701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henke, P.G. Hippocampal pathway to the amygdala and stress ulcer development. Brain Res. Bull. 1990, 25, 691–695. [Google Scholar] [CrossRef]
- Swanson, L.W.; Cowan, W.M. An autoradiographic study of the efferent connections of the entorhinal cortex in the rat. J. Comp. Neurol. 1977, 172, 49–84. [Google Scholar] [CrossRef]
- Thompson, C.L.; Pathak, S.D.; Jeromin, A.; Ng, L.L.; MacPherson, C.R.; Mortrud, M.T.; Cusick, A.; Riley, Z.L.; Sunkin, S.M.; Bernard, A.; et al. Genomic Anatomy of the Hippocampus. Neuron 2008, 60, 1010–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.W.; Swanson, L.W.; Chen, L.; Fanselow, M.S.; Toga, A.W. Genomic-anatomic evidence for distinct functional domains in hippocampal field CA1. Proc. Natl. Acad. Sci. USA 2009, 106, 11794–11799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anacker, C.; Hen, R. Adult hippocampal neurogenesis and cognitive flexibility-linking memory and mood. Nat. Rev. Neurosci. 2017, 18, 335–346. [Google Scholar] [CrossRef]
- Neves, G.; Cooke, S.F.; Bliss, T.V. Synaptic plasticity, memory and the hippocampus: A neural network approach to causality. Nat. Rev. Neurosci. 2012, 9, 1030–1034. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, J.T.; Schafer, S.T.; Gage, F.H. Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell 2016, 167, 897–914. [Google Scholar] [CrossRef] [Green Version]
- Gage, F.H. Adult neurogenesis in mammals. Science 2019, 364, 827–828. [Google Scholar] [CrossRef]
- Akers, K.G.; Martinez-Canabal, A.; Restivo, L.; Yiu, A.P.; De Cristofaro, A.; Hsiang, H.-L.; Wheeler, A.L.; Guskjolen, A.; Niibori, Y.; Shoji, H.; et al. Hippocampal Neurogenesis Regulates Forgetting During Adulthood and Infancy. Science 2014, 344, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Vivar, C.; Peterson, B.D.; van Praag, H. Running rewires the neuronal network of adult-born dentate granule cells. Neuroimage 2016, 131, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Van Praag, H.; Kempermann, G.; Gage, F.H. Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 1999, 2, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Bass, R.W.; Brown, D.D.; Laurson, K.R.; Coleman, M.M. Physical fitness and academic performance in middle school students. Acta Paediatr. Int. J. Paediatr. 2013, 102, 832–837. [Google Scholar] [CrossRef]
- Colcombe, S.J.; Erickson, K.I.; Raz, N.; Webb, A.G.; Cohen, N.J.; McAuley, E.; Kramer, A.F. Aerobic Fitness Reduces Brain Tissue Loss in Aging Humans. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2003, 58, M176–M180. [Google Scholar] [CrossRef] [Green Version]
- Pajonk, F.-G.; Wobrock, T.; Gruber, O.; Scherk, H.; Berner, D.; Kaizl, I.; Kierer, A.; Müller, S.; Oest, M.; Meyer, T.; et al. Hippocampal plasticity in response to exercise in schizophrenia. Arch. Gen. Psychiatry 2010, 67, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Phillips, C.; Baktir, M.A.; Srivatsan, M.; Salehi, A. Neuroprotective effects of physical activity on the brain: A closer look at trophic factor signaling. Front. Cell. Neurosci. 2014, 8, 1–16. [Google Scholar]
- Scharfman, H.; Goodman, J.; Macleod, A.; Phani, S.; Antonelli, C.; Croll, S. Increased neurogenesis and the ectopic granule cells after intrahippocampal BDNF infusion in adult rats. Exp. Neurol. 2005, 192, 348–356. [Google Scholar] [CrossRef]
- Åberg, M.A.I.; Åberg, N.D.; Hedbäcker, H.; Oscarsson, J.; Eriksson, P.S. Peripheral infusion of IGF-I selectively induces neurogenesis in the adult rat hippocampus. J. Neurosci. 2000, 20, 2896–2903. [Google Scholar] [CrossRef] [Green Version]
- Gleeson, J.G.; Lin, P.T.; Flanagan, L.A.; Walsh, C.A. Doublecortin Is a Microtubule-Associated Protein and Is Expressed Widely by Migrating Neurons Neurons migrating either along radial glia or indepen- dent of radial glia demonstrate a variety of cytoskeletal changes within the soma and processes that may u. Neuron 1999, 23, 257–271. [Google Scholar] [CrossRef] [Green Version]
- Couillard-Despres, S.; Winner, B.; Schaubeck, S.; Aigner, R.; Vroemen, M.; Weidner, N.; Bogdahn, U.; Winkler, J.; Kuhn, H.G.; Aigner, L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Toyama, B.H.; Harris, M.S.; Bock, T.; Iskar, M.; Bork, P.; Ingolia, N.T.; Hetzer, M.W.; Beck, M. Integrated Transcriptome and Proteome Analyses Reveal Organ-Specific Proteome Deterioration in Old Rats. Cell Syst. 2015, 1, 224–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duda, P.; Wójcicka, O.; Wisniewski, J.R.; Rakus, D. Global quantitative TPA-based proteomics of mouse brain structures reveals significant alterations in expression of proteins involved in neuronal plasticity during aging. Aging 2018, 10, 1682–1697. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Lozano, M.A.; Klemmer, P.; Gebuis, T.; Hassan, C.; Van Nierop, P.; Van Kesteren, R.E.; Smit, A.B.; Li, K.W. Dynamics of the mouse brain cortical synaptic proteome during postnatal brain development. Sci. Rep. 2016, 6, 35456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers - Assembly, dynamics and biological roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, J.S.; Medina, M.; Zuliani, C.; Di Nardo, A.; Witke, W.; Dotti, C.G. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J. Cell Biol. 2003, 162, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
- Colgan, L.A.; Yasuda, R. Plasticity of dendritic spines: Subcompartmentalization of signaling. Annu. Rev. Physiol. 2014, 76, 365–385. [Google Scholar] [CrossRef] [Green Version]
- Boles, N.C.; Hirsch, S.E.; Le, S.; Corneo, B.; Najm, F.; Minotti, A.P.; Wang, Q.; Lotz, S.; Tesar, P.J.; Fasano, C.A. NPTX1 Regulates Neural Lineage Specification from Human Pluripotent Stem Cells. Cell Rep. 2014, 6, 724–736. [Google Scholar] [CrossRef] [Green Version]
- Tobin, M.K.; Musaraca, K.; Disouky, A.; Shetti, A.; Bheri, A.; Honer, W.G.; Kim, N.; Dawe, R.J.; Bennett, D.A.; Arfanakis, K.; et al. Human Hippocampal Neurogenesis Persists in Aged Adults and Alzheimer’s Disease Patients. Cell Stem Cell 2019, 24, 974–982. [Google Scholar] [CrossRef]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef]
- Horgusluoglu, E.; Nudelman, K.; Nho, K.; Saykin, A.J. Adult neurogenesis and neurodegenerative diseases: A systems biology perspective. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2017, 174, 93–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lourenco, M.V.; Frozza, R.L.; de Freitas, G.B.; Zhang, H.; Kincheski, G.C.; Ribeiro, F.C.; Gonçalves, R.A.; Clarke, J.R.; Beckman, D.; Staniszewski, A.; et al. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer’s models. Nat. Med. 2019, 25, 165–175. [Google Scholar] [CrossRef]
- Wei, R.; Wang, J.; Su, M.; Jia, E.; Chen, S.; Chen, T.; Ni, Y. Missing Value Imputation Approach for Mass Spectrometry-based Metabolomics Data. Sci. Rep. 2018, 8, 663. [Google Scholar] [CrossRef] [Green Version]
- Tanti, A.; Westphal, W.P.; Girault, V.; Brizard, B.; Devers, S.; Leguisquet, A.M.; Surget, A.; Belzung, C. Region-dependent and stage-specific effects of stress, environmental enrichment, and antidepressant treatment on hippocampal neurogenesis. Hippocampus 2013, 23, 797–811. [Google Scholar] [CrossRef] [PubMed]
- Tanti, A.; Rainer, Q.; Minier, F.; Surget, A.; Belzung, C. Differential environmental regulation of neurogenesis along the septo-temporal axis of the hippocampus. Neuropharmacology 2012, 63, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Farris, S.; Ward, J.M.; Carstens, K.E.; Samadi, M.; Wang, Y.; Dudek, S.M. Hippocampal Subregions Express Distinct Dendritic Transcriptomes that Reveal Differences in Mitochondrial Function in CA2. Cell Rep. 2019, 29, 522–539. [Google Scholar] [CrossRef] [Green Version]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260. [Google Scholar] [CrossRef] [Green Version]
- Black, J.E.; Isaacs, K.R.; Anderson, B.J.; Alcantara, A.A.; Greenough, W.T. Learning causes synaptogenesis, whereas motor activity causes angiogenesis, in cerebellar cortex of adult rats. Proc. Natl. Acad. Sci. USA 1990, 87, 5568–5572. [Google Scholar] [CrossRef] [Green Version]
- Merikangas, K.R.; Swendsen, J.; Hickie, I.B.; Cui, L.; Shou, H.; Merikangas, A.K.; Zhang, J.; Lamers, F.; Crainiceanu, C.; Volkow, N.D.; et al. Real-time Mobile Monitoring of the Dynamic Associations among Motor Activity, Energy, Mood, and Sleep in Adults with Bipolar Disorder. JAMA Psychiatry 2019, 76, 190–198. [Google Scholar] [CrossRef]
- Follwaczny, P.; Schieweck, R.; Riedemann, T.; Demleitner, A.; Straub, T.; Klemm, A.H.; Bilban, M.; Sutor, B.; Popper, B.; Kiebler, M.A. Pumilio2-deficient mice show a predisposition for epilepsy. Dis. Model. Mech. 2017, 10, 1333–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popper, B.; Demleitner, A.; Bolivar, V.J.; Kusek, G.; Snyder-Keller, A.; Schieweck, R.; Temple, S.; Kiebler, M.A. Staufen2 deficiency leads to impaired response to novelty in mice. Neurobiol. Learn. Mem. 2018, 150, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Kheirbek, M.A.; Drew, L.J.; Burghardt, N.S.; Costantini, D.O.; Tannenholz, L.; Ahmari, S.E.; Zeng, H.; Fenton, A.A.; Henl, R. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron 2013, 77, 955–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzózka, M.M.; Havemann-Reinecke, U.; Wichert, S.P.; Falkai, P.; Rossner, M.J. Molecular signatures of psychosocial stress and cognition are modulated by chronic lithium treatment. Schizophr. Bull. 2016, 42, S22–S33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frey, S.; Schieweck, R.; Forné, I.; Imhof, A.; Straub, T.; Popper, B.; Kiebler, M.A. Physical Activity Dynamically Regulates the Hippocampal Proteome along the Dorso-Ventral Axis. Int. J. Mol. Sci. 2020, 21, 3501. https://doi.org/10.3390/ijms21103501
Frey S, Schieweck R, Forné I, Imhof A, Straub T, Popper B, Kiebler MA. Physical Activity Dynamically Regulates the Hippocampal Proteome along the Dorso-Ventral Axis. International Journal of Molecular Sciences. 2020; 21(10):3501. https://doi.org/10.3390/ijms21103501
Chicago/Turabian StyleFrey, Surina, Rico Schieweck, Ignasi Forné, Axel Imhof, Tobias Straub, Bastian Popper, and Michael A. Kiebler. 2020. "Physical Activity Dynamically Regulates the Hippocampal Proteome along the Dorso-Ventral Axis" International Journal of Molecular Sciences 21, no. 10: 3501. https://doi.org/10.3390/ijms21103501




