Small RNA Sequencing Analysis of miRNA Expression Reveals Novel Insihts into Root Formation under Root Restriction Cultivation in Grapevine (Vitis vinifera L.)
Abstract
:1. Introduction
2. Results
2.1. Phenotype Variations of Grapevine cv. Muscat Hamburg after Root Restriction Cultivation
2.2. Sequencing Statistics in Different Grapevine Samples
2.3. Identification of Known and Novel Grapevine miRNAs
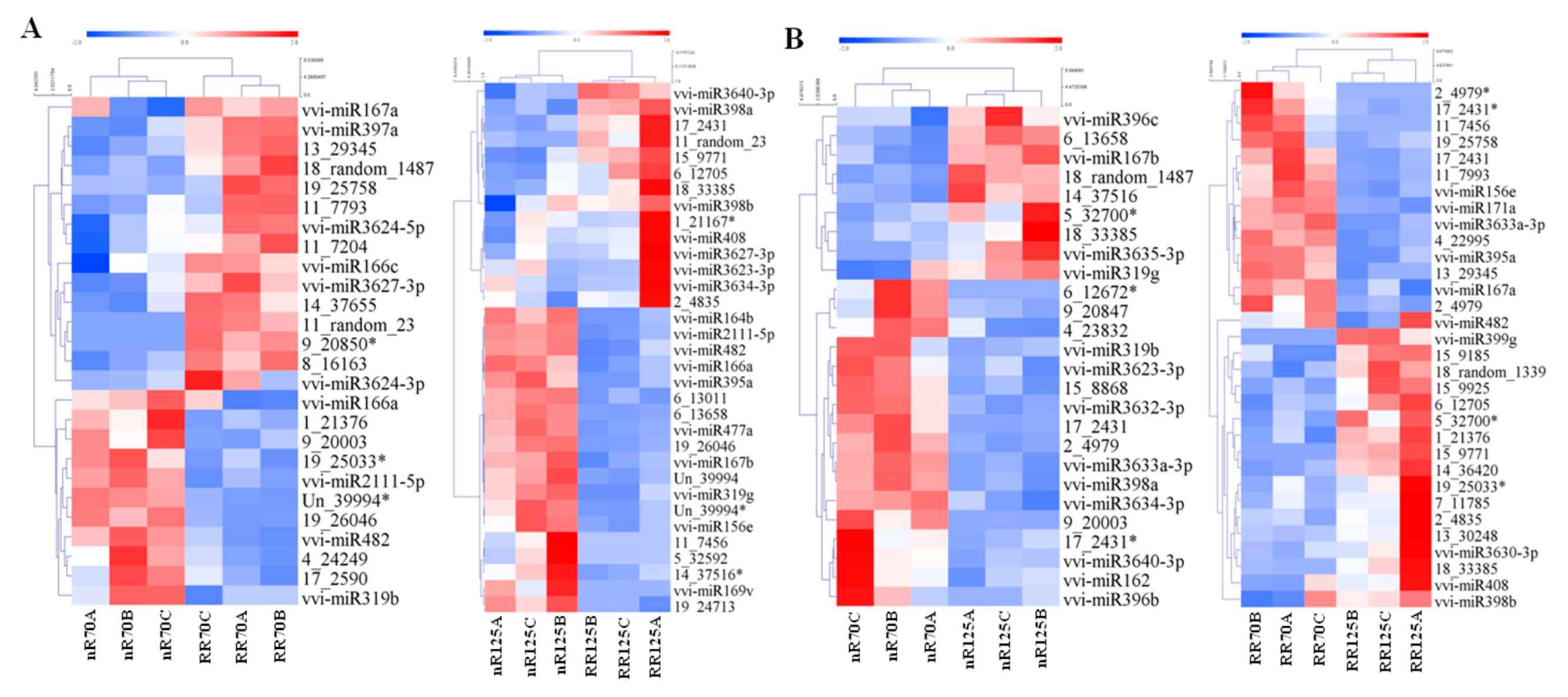
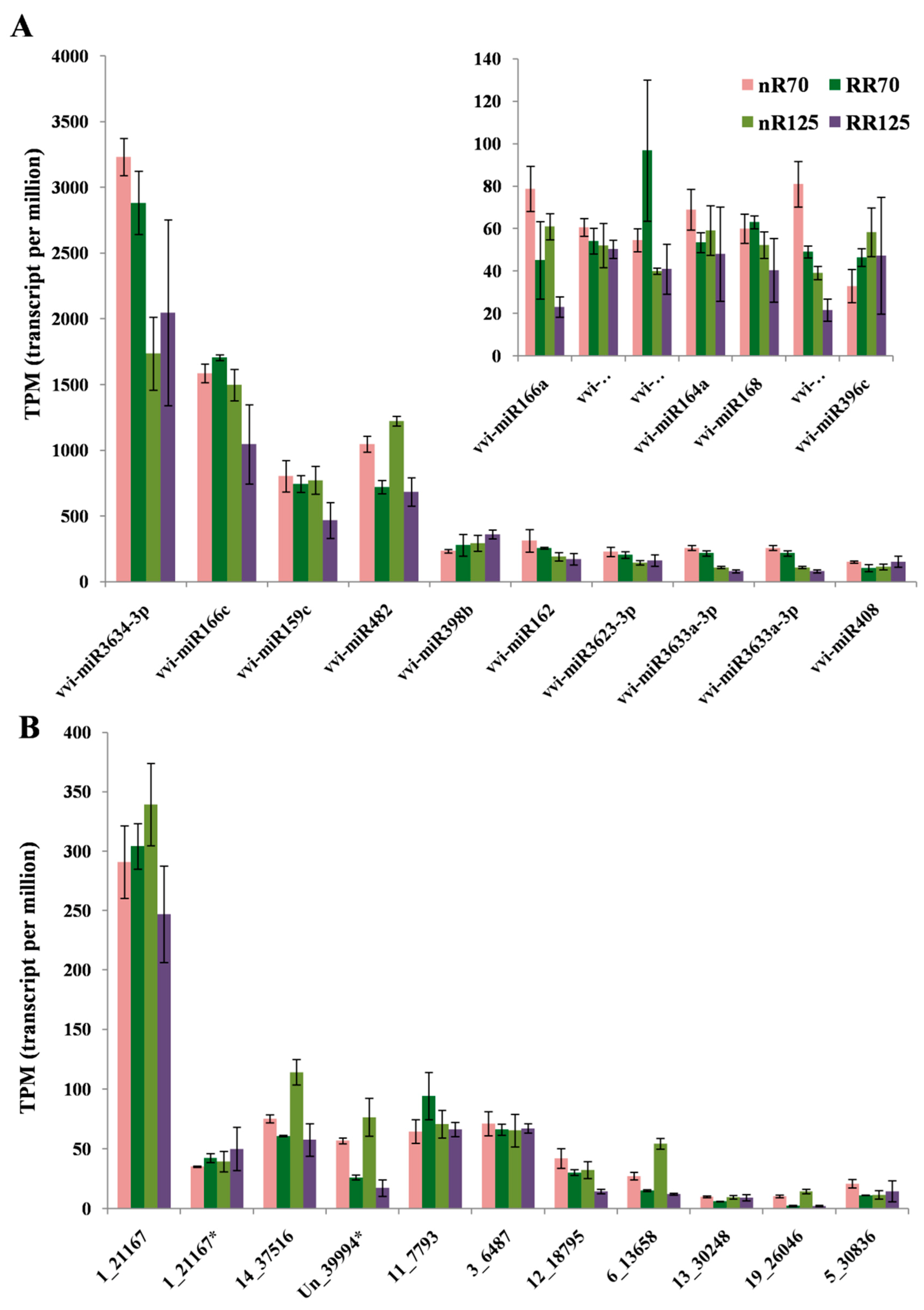
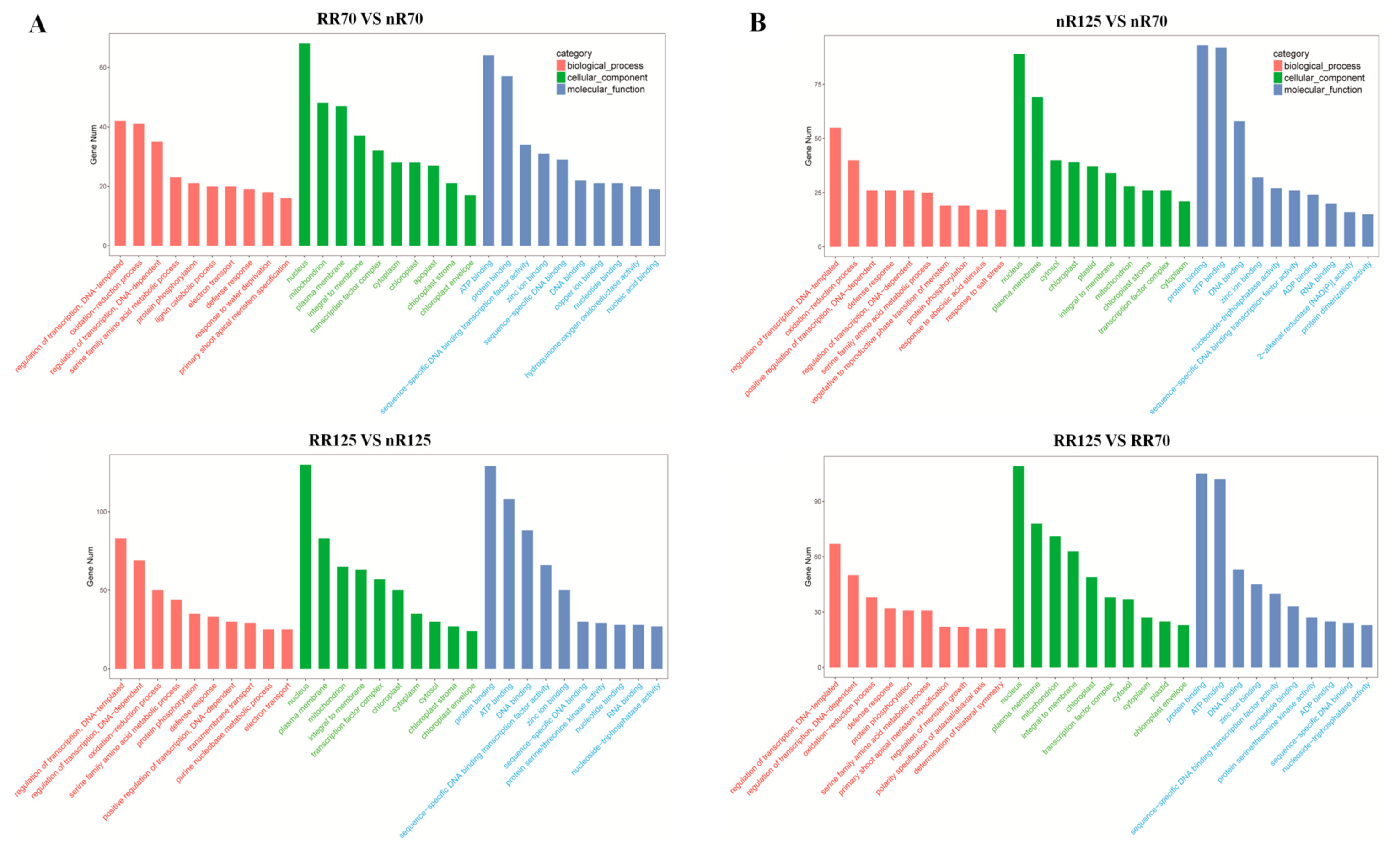
2.4. Differentially Expressed miRNAs (DEMs) Analysis
2.5. Analysis of vvi-miRNA Mediated Grapevine Root Formation
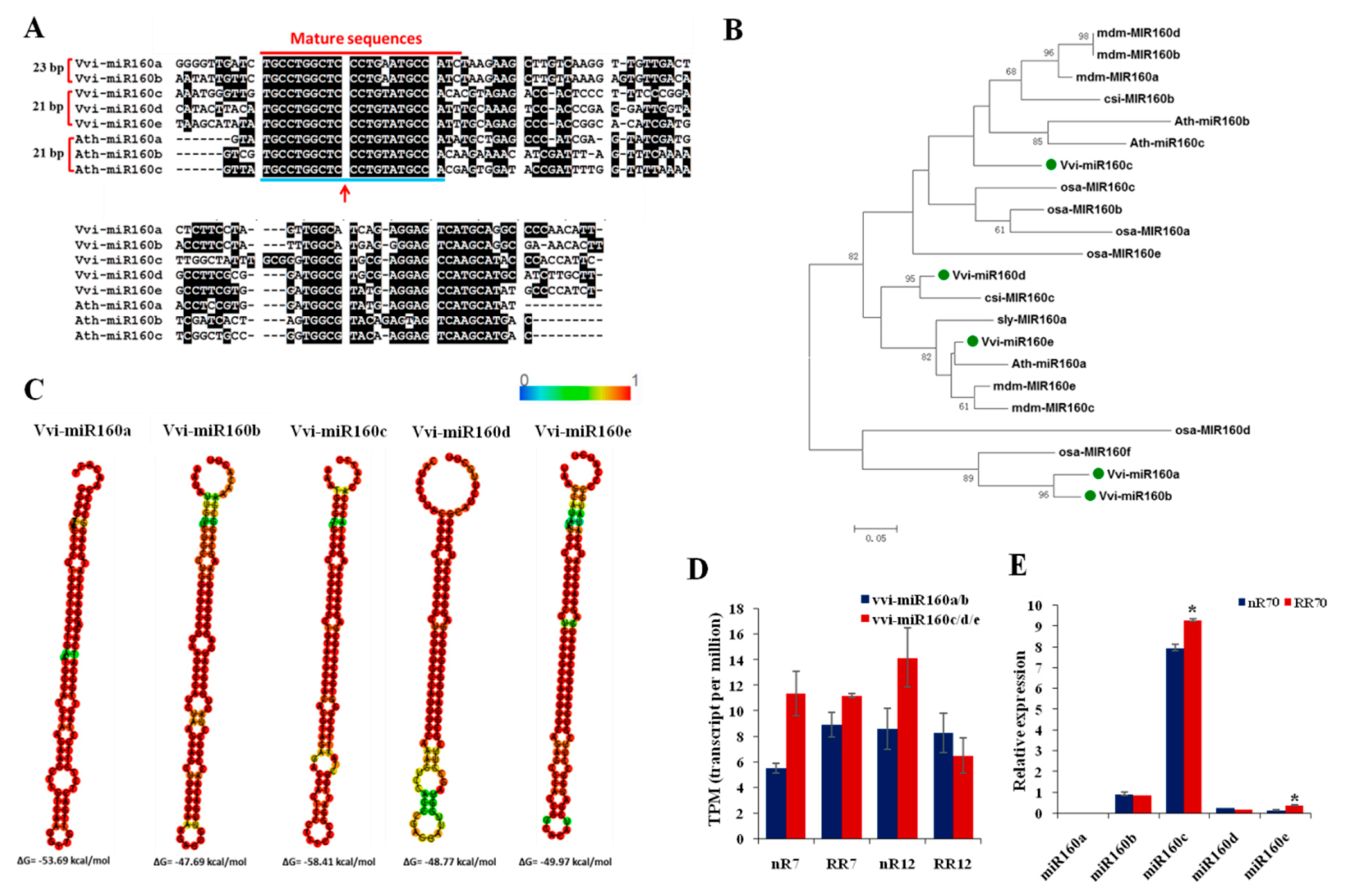
2.6. Vvi-miR160 Family Contributes to Grapevine Root Development
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Small RNA Libraries Construction and Illumina Sequencing
4.3. Identification of Known and Novel vvi-miRNAs
4.4. Differentially Expressed miRNAs Analysis and Annotation of the Target Genes
4.5. Structure Analysis of vvi-miR160 Family
4.6. RT-qPCR Analysis of the Expression Levels of vvi-miR160 Family
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ARF | Auxin response factor |
| bHLH | basic helix-loop-helix |
| DAP | Days after planting |
| DEMs | Differentially expressed miRNAs |
| miRNA | microRNA |
| NCBI | the National Centre for Biotechnology Information |
| ncRNA | Non-coding RNA |
| nRC | Non-Root restriction cultivation |
| PCA | Principal component analysis |
| RRC | Root restriction cultivation |
| Spl | Squamosa-promoter binding protein-like |
| TAS | Trans-acting siRNA genes |
| TPM | Transcript per million |
References
- Tardieu, F. Drought perception by plants do cells of droughted plants experience water stress? Plant Growth Regul. 1996, 20, 93–104. [Google Scholar] [CrossRef]
- Hopmans, J.W.; Bristow, K.L. Current capabilities and future needs of root water and nutrient uptake modeling. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2002; Volume 77, pp. 103–183. [Google Scholar]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voss, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching Out in Roots: Uncovering Form, Function, and Regulation. Plant Physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ingram, P.A.; Malamy, J.E. Root system architecture. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2010; Volume 55, pp. 75–117. [Google Scholar]
- Riaz, S.; Garrison, K.E.; Dangl, G.S.; Boursiquot, J.-M.; Meredith, C.P. Genetic divergence and chimerism within ancient asexually propagated winegrape cultivars. J. Am. Soc. Hortic. Sci. 2002, 127, 508–514. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Gao, Z.; Chen, Q.; Li, Q.; Luo, M.; Wang, J.; Hu, L.; Zahid, M.S.; Wang, L.; Zhao, L.; et al. Grapevine ABA receptor VvPYL1 regulates root hair development in Transgenic Arabidopsis. Plant Physiol. Biochem. PPB 2020, 149, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Okamoto, G.; Hirano, K.; Lu, J.; Zhang, C. Effects of restricted rooting volume on vine growth and berry development of Kyoho grapevines. Am. J. Enol. Vitic. 2001, 52, 248–253. [Google Scholar]
- Naor, A.; Gal, Y.; Bravdo, B. Shoot and cluster thinning influence vegetative growth, fruit yield, and wine quality of Sauvignon blanc’ grapevines. J. Am. Soc. Hortic. Sci. 2002, 127, 628–634. [Google Scholar] [CrossRef] [Green Version]
- Leng, F.; Lin, Q.; Wu, D.; Wang, S.; Wang, D.; Sun, C. Comparative transcriptomic analysis of grape berry in response to root restriction during developmental stages. Molecules 2016, 21, 1431. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Okamoto, G.; Hirano, K. Effects of rooting-zone restriction on the changes in carbohydrates and nitrogenous compounds in ‘Kyoho’ grapevines during winter dormancy and early shoot growth. J. Jpn. Soc. Hortic. Sci. 1998, 67, 577–582. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.-M.; Li, J.-F.; Zhu, L.-N.; Wang, B.; Wang, L.; Bai, Y.; ZHANG, C.-X.; Xu, W.-P.; Wang, S.-P. Effects of root restriction on nitrogen and gene expression levels in nitrogen metabolism in Jumeigui grapevines (Vitis vinifera L.× Vitis labrusca L.). J. Integr. Agric. 2015, 14, 67–79. [Google Scholar] [CrossRef]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol. A J. Pathol. Soc. Great Br. Irel. 2010, 220, 126–139. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet. 2006, 15 (Suppl. 1), R17–R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 2013, 153, 562–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, Y.; Ma, X.; Chen, D.; Wu, P.; Chen, M. MicroRNA-mediated signaling involved in plant root development. Biochem. Biophys. Res. Commun. 2010, 393, 345–349. [Google Scholar] [CrossRef]
- Meng, Y.; Wu, P.; Chen, M. MicroRNAs in plant roots: Current understanding and future perspectives. In Non Coding RNAs in Plants; Springer: Berlin/Heidelberg, Germany, 2011; pp. 269–284. [Google Scholar]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef] [Green Version]
- Bustos-Sanmamed, P.; Mao, G.; Deng, Y.; Elouet, M.; Khan, G.A.; Bazin, J.; Turner, M.; Subramanian, S.; Yu, O.; Crespi, M. Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 2013, 40, 1208–1220. [Google Scholar] [CrossRef]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [Green Version]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–236. [Google Scholar] [CrossRef]
- Snyman, M.C.; Solofoharivelo, M.-C.; Souza-Richards, R.; Stephan, D.; Murray, S.; Burger, J.T. The use of high-throughput small RNA sequencing reveals differentially expressed microRNAs in response to aster yellows phytoplasma-infection in Vitis vinifera cv. ‘Chardonnay’. PLoS ONE 2017, 12, e0182629. [Google Scholar] [CrossRef]
- Sun, X.; Fan, G.; Su, L.; Wang, W.; Liang, Z.; Li, S.; Xin, H. Identification of cold-inducible microRNAs in grapevine. Front. Plant Sci. 2015, 6, 595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullan, J.B.; Pinto, D.L.P.; Bertolini, E.; Fasoli, M.; Zenoni, S.; Tornielli, G.B.; Pezzotti, M.; Meyers, B.C.; Farina, L.; Pè, M.E. miRVine: A microRNA expression atlas of grapevine based on small RNA sequencing. BMC Genom. 2015, 16, 393. [Google Scholar]
- Mica, E.; Piccolo, V.; Delledonne, M.; Ferrarini, A.; Pezzotti, M.; Casati, C.; Del Fabbro, C.; Valle, G.; Policriti, A.; Morgante, M. High throughput approaches reveal splicing of primary microRNA transcripts and tissue specific expression of mature microRNAs in Vitis vinifera. BMC Genom. 2009, 10, 558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitarra, W.; Pagliarani, C.; Abbà, S.; Boccacci, P.; Birello, G.; Rossi, M.; Palmano, S.; Marzachì, C.; Perrone, I.; Gambino, G. miRVIT: A novel miRNA database and its application to uncover vitis responses to flavescence dorée infection. Front. Plant Sci. 2018, 9, 1034. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Deng, B.; Gao, J.; Zhao, Z.; Chen, Z.; Song, S.; Wang, L.; Zhao, L.; Xu, W.; Zhang, C. Comparative Analysis of miRNA Abundance Revealed the Function of Vvi-miR828 in Fruit Coloring in Root Restriction Cultivation Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2019, 20, 4058. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. miR396a-mediated basic helix–loop–helix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Peng, S.; Xian, Z.; Lin, D.; Hu, G.; Yang, L.; Ren, M.; Li, Z. Overexpression of a tomato miR171 target gene SlGRAS 24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol. J. 2017, 15, 472–488. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic Plasticity of Adventitious Rooting in Arabidopsis is Controlled by Complex Regulation of AUXIN RESPONSE FACTOR Transcripts and MicroRNA Abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Q.-H.; Fan, L.; Liu, Y.; Xu, H.; Llewellyn, D.; Wilson, I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE 2013, 8, e84390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, V.M.; Müller, S.; Baulcombe, D.; Puigdomènech, P. Evolution of NBS-LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Mol. Plant 2015, 8, 329–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Galili, G. Tuning the Orchestra: miRNAs in Plant Immunity. Trends Plant Sci. 2019, 24, 189–191. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [Green Version]
- Ni, Z.; Hu, Z.; Jiang, Q.; Zhang, H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef]
- Bakhshi, B.; Fard, E.M.; Hosseini, G. Evaluation of miR398 differential expression in rice under drought stress condition. Bull. Georg. Natl. Acad. Sci 2014, 8, 87–92. [Google Scholar]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Gordon, A.; Hannon, G. Fastx-toolkit. FASTQ/A Short-Reads Preprocessing Tools. Unpublished. May 2010. Available online: http://hannonlab.cshl.edu/fastx_toolkit (accessed on 5 April 2020).
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Bateman, A.; Marshall, M.; Khanna, A.; Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003, 31, 439–441. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 25, 4.10.1–4.10.14. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36 (Suppl. 1), D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Denman, R.B. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques 1993, 15, 1090–1095. [Google Scholar] [PubMed]
- Sun, J.; Wang, S.; Li, C.; Ren, Y.; Wang, J. Novel expression profiles of microRNAs suggest that specific miRNAs regulate gene expression for the sexual maturation of female Schistosoma japonicum after pairing. Parasites Vectors 2014, 7, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S. Analysing RNA-Seq data with the DESeq package. Mol. Biol. 2010, 43, 1–17. [Google Scholar]
- Fahlgren, N.; Carrington, J.C. miRNA target prediction in plants. In Plant MicroRNAs; Springer: Berlin/Heidelberg, Germany, 2010; pp. 51–57. [Google Scholar]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Luo, M.; Gao, Z.; Li, H.; Li, Q.; Zhang, C.; Xu, W.; Song, S.; Ma, C.; Wang, S. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine. Sci. Rep. 2018, 8, 4444. [Google Scholar] [CrossRef]




| Sample | Raw Reads | Reads Trimmed Length | Reads Trimmed_Q20 | Reads Trimmed_N | Clean Reads | Clean Reads Uniq |
|---|---|---|---|---|---|---|
| nR70A | 17,440,227 | 13,828,456 | 13,805,784 | 13,791,775 | 13,791,775 | 2,789,195 |
| nR70B | 16,785,530 | 12,712,507 | 12,694,953 | 12,682,543 | 12,682,543 | 2,424,073 |
| nR70C | 16,705,937 | 12,741,508 | 12,719,806 | 12,706,889 | 12,706,889 | 2,449,057 |
| RR70A | 19,603,314 | 16,149,312 | 16,130,501 | 16,129,531 | 16,129,531 | 3,896,588 |
| RR70B | 16,454,204 | 13,692,986 | 13,676,330 | 13,662,899 | 13,662,899 | 3,129,670 |
| RR70C | 14,509,081 | 11,845,746 | 11,840,120 | 11,839,622 | 11,839,622 | 2,768,680 |
| nR125A | 17,921,125 | 14,407,233 | 14,391,593 | 14,377,909 | 14,377,909 | 3,583,714 |
| nR125B | 18,875,954 | 14,074,226 | 14,050,637 | 14,036,500 | 14,036,500 | 3,639,479 |
| nR125C | 17,047,738 | 12,015,900 | 11,997,503 | 11,985,776 | 11,985,776 | 3,224,084 |
| RR125A | 19,152,378 | 15,408,723 | 15,401,439 | 15,400,735 | 15,400,735 | 3,942,147 |
| RR125B | 20,246,983 | 16,368,921 | 16,361,306 | 16,360,589 | 16,360,589 | 4,167,784 |
| RR125C | 19,697,117 | 15,784,039 | 15,767,917 | 15,766,919 | 15,766,919 | 4,360,497 |
| Total | 214,439,588 | 169,029,557 | 168,837,889 | 168,741,687 | 168,741,687 | 40,374,968 |
| Provisional ID | Mature Sequence | Length | Loc_miRNA | miRBase Alignment | Query Alignment | Subject Alignment | Strand | Score | Evalue |
|---|---|---|---|---|---|---|---|---|---|
| 6_12672 | AGCUGCCGACUCAUUCAUUCA | 21 | chr6:9137310_9137389:+ | csi-miR159b-5p | 1_21 | 1_21 | + | 105 | 0.002 |
| 17_2431 | AGCUGCUGACUUAUGGAUCCC | 21 | chr17:2609257_2609342:- | mes-miR159a-5p | 1_21 | 1_21 | + | 87 | 0.065 |
| 9_19848 | CUCUCUGCUACCGUCAUUCUGC | 22 | chr9:19106874_19106939:+ | csi-miR156f-3p | 1_18 | 4_21 | + | 63 | 7 |
| 15_8904 | UCGAGAAACCUCUGCAUCA | 19 | chr15:9049770_9049811:+ | ath-miR162a-3p | 1_18 | 1_18 | + | 81 | 0.2 |
| 2_4979 | AUUGAAUGAUGCGGGAGACA | 20 | chr2:855616_855687:- | seu-miR319 | 3_20 | 3_20 | + | 81 | 0.22 |
| 11_7793 | UCCCACAGCUUUCUUGAACUU | 21 | chr11:5246803_5246886:- | vvi-miR396b | 1_20 | 1_20 | + | 91 | 0.03 |
| 9_20339 | CUUUCUUGAACCAAUGGGUCCCAUU | 25 | chr9:5984685_5984768:- | osa-miR396e-3p | 1_13 | 2_14 | - | 65 | 4.3 |
| 19_26046 | GUUGGAAGUCGGUGGGGGACC | 21 | chr19:18872730_18872799:- | ppt-miR477f | 1_18 | 1_18 | + | 90 | 0.037 |
| 19_25033 | UCCCUCAAAGGCUUCCAAUUU | 21 | chr19:18678362_18678430:+ | ppt-miR477f | 1_19 | 1_18 | + | 90 | 0.037 |
| 6_13658 | CUGGAAGCCGAUGGGGGACC | 20 | chr6:19950754_19950822:- | ppt-miR477f | 1_20 | 1_18 | + | 90 | 0.037 |
| Un_39994 | UCCCUCAAAGGCUUCCAAUUUU | 22 | chrUn:16672987_16673055:- | ppt-miR477f | 1_21 | 1_18 | + | 90 | 0.04 |
| 19_26048 | AAGUUGGAAGCCGGUGGGGGA | 21 | chr19:18881377_18881443:- | ppt-miR477f | 3_21 | 1_19 | - | 68 | 2.5 |
| 14_37516 | UUCCCAAUGCCGCCCAUUCCAA | 22 | chr14:19755490_19755563:- | gma-miR482a-3p | 1_22 | 3_24 | + | 92 | 0.027 |
| 1_21167 | UCUUACCAACACCUCCCAUUCCA | 23 | chr1:3865584_3865662:+ | mtr-miR482-3p | 1_22 | 1_21 | + | 87 | 0.065 |
| MiRNA_id | Target Gene’ mRNA | Target Start | Target End | Strand | Score | Target Seq (5′->3′) | Query Seq (3′->5′) | GO id | Gene Function Annoation | KEGG Pathway | KEGG Pathway Description |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4_24249 | VIT_214s0068g01330 | 3178 | 3196 | + | 4 | AAACGGAAGAACCACCAUA | AUAGCCUUAUUGGUGGUAU | GO:0080022 | primary root development | NA | NA |
| 17_2431 | VIT_212s0057g00680 | 1326 | 1346 | + | 4 | GGAGUUCCUGAGUCAGCAGCU | CCCUAGGUAUUCAGUCGUCGA | GO:0080022; GO:0010071; GO:0010078 | primary root development |root meristem specification | maintenance of root meristem identity | NA | NA |
| 15_8868 > 15_8867 | VIT_208s0058g01340 | 2392 | 2409 | + | 3 | CAUGCUGCUGCCAGCCCA | CCACCACGACGGUCGGGU | GO:0016021 | root morphogenesis | NA | NA |
| 15_8868 > 15_8867 | VIT_211s0037g00040 | 1169 | 1185 | + | 4 | GGUGGUGCU-CCAGCACA | CCACCACGACGGUCGGGU | GO:0080147 | root hair cell development | NA | NA |
| 6_12672* | VIT_203s0063g02440 | 545 | 562 | + | 4 | ACC-CCCGUAUUAUUCAUA | UGGAGGGCGUAGUAAGUGU | GO:0048364 | root development | NA | NA |
| 6_13011 | VIT_211s0016g01900 | 119 | 136 | + | 4 | AAGUCAUCGCA-CACAGCA | UUCAGUAGAGUAGUGUCGU | GO:0048528 | post-embryonic root development | NA | NA |
| 9_20003 | VIT_214s0066g00370 | 1930 | 1947 | + | 4 | GCUGUCUUG-CCACCAGAG | UGGCGGAACUGGUGGUCUU | GO:0010053 | root epidermal cell differentiation | NA | NA |
| 9_20003 | VIT_214s0066g00370 | 1930 | 1947 | + | 4 | GCUGUCUUG-CCACCAGAG | UGGCGGAACUGGUGGUCUU | GO:0010053 | root epidermal cell differentiation | NA | NA |
| 19_25758 | VIT_201s0010g03300 | 3037 | 3054 | + | 4 | GAGGCAUCCGUAUUUUCA | CUCGGUAGGCGUCAAAGU | GO:0010053 | root epidermal cell differentiation | ko00510, ko04141 | N-Glycan biosynthesis | Protein processing in endoplasmic reticulum |
| 19_24713 | VIT_213s0019g02390 | 1712 | 1728 | + | 4 | UCAUUCA-CUCAAAAUCU | AGUCAGUUGGGUUUUAGA | GO:0048767; GO:0010449 | root hair elongation | root meristem growth | ko00790 | Folate biosynthesis |
| 19_24713 | VIT_213s0019g04320 | 129 | 146 | + | 4 | UUGUUCAACCCCAAAUCU | AGUCAGUUGGGUUUUAGA | GO:0048765 | root hair cell differentiation | NA | NA |
| 19_24713 | VIT_203s0038g03620 | 4817 | 4833 | + | 4 | UCAG-CAACCAAAAAUCA | AGUCAGUUGGGUUUUAGA | GO:0048767 | root hair elongation | NA | NA |
| vvi-miR3624-3p | VIT_205s0029g00330 | 1110 | 1130 | + | 4 | AGGGCUGCGCUGCUGCCCUGA | UCAUCAUACGACGACGGGACU | GO:0005829 | root hair elongation | ko00270 | Cysteine and methionine metabolism |
| 2_4979* | VIT_218s0001g04980 | 889 | 907 | + | 4 | GGAUGAAUGAGUCGG-AGAU | CCUACUUACUCAGCCGUCGA | GO:0048364 | root development | ko01212, ko00620, ko00640, ko00061, ko00254, ko04152 | Fatty acid metabolism | Pyruvate metabolism | Propanoate metabolism | Fatty acid biosynthesis | Aflatoxin biosynthesis | AMPK signaling pathway |
| 2_4979* | VIT_207s0289g00100 | 395 | 413 | + | 4 | GGAUGAAUGAGUCGG-AGAU | CCUACUUACUCAGCCGUCGA | GO:0048364 | root development | NA | NA |
| vvi-miR396 | VIT_208s0007g03760 | 703 | 723 | + | 3 | CGUUCAAGAAAGCCUGUGGAA | UCAAGUUCUUUCG-ACACCUU | GO:0048364 | root development | NA | NA |
| vvi-miR2111-5p | VIT_208s0007g01270 | 1635 | 1655 | + | 0 | UAGACCUCAGGAUGCAGAUUA | AUCUGGAGUCCUACGUCUAAU | GO:0080147 | root hair cell development | ko04962 | Vasopressin-regulated water re-absorption |
| vvi-miR156 | VIT_204s0008g00960 | 2493 | 2512 | + | 1 | GUGCUCACUCUCUUCUGUCA | CACGAGUGAGAGGAGACAGU | GO:0048767 | root hair elongation | ko04010, ko04020 | MAPK signaling pathway | Calcium signaling pathway |
| vvi-miR166 | VIT_209s0002g03740 | 1510 | 1528 | + | 0.5 | GGAAUGAAGCCUGGUCCGG | CCUUACUUCGGACCAGGCU | GO:0048765 | root hair cell differentiation | NA | NA |
| vvi-miR166 | VIT_206s0004g02800 | 1730 | 1748 | + | 1 | GGGAUGAAGCCUGGUCCGG | CCUUACUUCGGACCAGGCU | GO:0045595 | root hair cell differentiation | NA | NA |
| vvi-miR166 | VIT_213s0019g04320 | 1114 | 1132 | + | 1 | GGGAUGAAGCCUGGUCCGG | CCUUACUUCGGACCAGGCU | GO:0008289 | root hair cell differentiation | NA | NA |
| vvi-miR166 | VIT_209s0002g03740 | 1508 | 1528 | + | 2.5 | CUGGAAUGAAGCCUGGUCCGG | CCCCUUACUUCGGACCAGGCU | GO:0048263 | root hair cell differentiation | NA | NA |
| vvi-miR164 | VIT_219s0027g00230 | 589 | 609 | + | 1.5 | AGCAAGUGCCCUGCUUCUCCG | UCGUACACGGGACGAAGAGGU | GO:0048527 | lateral root development | NA | NA |
| vvi-miR482 | VIT_218s0001g03540 | 372 | 392 | + | 4 | GGAGUGAGAGGAG-AGGAAAGG | CCUUACCCUCCUCAUCCUUUCU | GO:0010311; GO:0048829 | lateral root formation | root cap development | ko04075 | Plant hormone signal transduction |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Gao, Z.; Zahid, M.S.; Li, D.; Javed, H.U.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Small RNA Sequencing Analysis of miRNA Expression Reveals Novel Insihts into Root Formation under Root Restriction Cultivation in Grapevine (Vitis vinifera L.). Int. J. Mol. Sci. 2020, 21, 3513. https://doi.org/10.3390/ijms21103513
Li H, Gao Z, Zahid MS, Li D, Javed HU, Wang L, Song S, Zhao L, Xu W, Zhang C, et al. Small RNA Sequencing Analysis of miRNA Expression Reveals Novel Insihts into Root Formation under Root Restriction Cultivation in Grapevine (Vitis vinifera L.). International Journal of Molecular Sciences. 2020; 21(10):3513. https://doi.org/10.3390/ijms21103513
Chicago/Turabian StyleLi, Hui, Zhen Gao, Muhammad Salman Zahid, Dongmei Li, Hafiz Umer Javed, Lei Wang, Shiren Song, Liping Zhao, Wenping Xu, Caixi Zhang, and et al. 2020. "Small RNA Sequencing Analysis of miRNA Expression Reveals Novel Insihts into Root Formation under Root Restriction Cultivation in Grapevine (Vitis vinifera L.)" International Journal of Molecular Sciences 21, no. 10: 3513. https://doi.org/10.3390/ijms21103513




