Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression
Abstract
1. Introduction
2. Mechanisms of AGEs-RAGE Axis Cancer Progression
2.1. ChREBP
2.2. JAK/STAT3
2.3. MAPK and MMPs
2.4. Nrf-2
2.5. PI3K/Akt
| Cell Lines | Study Type/Samples | Mechanisms of Action | References |
|---|---|---|---|
| Human colon adenocarcinoma (Caco-2) | In vitro | ↑ERK1/2, ↑MAPK | [27] |
| Human colon carcinoma (Colo320 and WiDr) | In vitro | ↑NF-κB, ↑ERK | [13] |
| Human erythroleukemia (HEL) | In vitro | ↑MAPK, ↑PI3K, ↑JAK/STAT | [33] |
| Human breast cancer (MCF-7) | In vitro | ↑VEGF | [24] |
| Human breast tumor samples | In vivo | ↑NF-κB | [42] |
| Human hepatocellular carcinoma (Hep3B and HepG2) | In vitro | ↑VEGF | [21] |
| Glioma (C6) | In vitro | ↑NF-κB | [43] |
| Human colorectal carcinoma (HCT116) | In vitro | ↑ChREBP | [28] |
| Human hepatocellular carcinoma (HepG2) | In vitro | ↑ChREBP | [28] |
| Human oral cancer (SAS) | In vitro | ↑ERK, ↑MMP2, ↑MMP9 | [39] |
| Human prostate cancer (PC-3) | In vitro | ↑PI3K/Akt | [22] |
| Human breast cancer (MDA-MB-231) | In vitro | ↑ERK1/2, ↑STAT3, ↑p38, ↑MAPK, ↑MMP-9 | [25] |
| Human gastric cancer (SGC7901) and human gastric tumors samples | In vitro/ In vivo | ↑ERK/Sp1/MMP2 | [34] |
| Rat pheochromocytoma (PC12) | In vitro | ↑NF-κB | [44] |
| Human breast cancer (MCF-7) | In vitro | ↑MMP9, ↑ERK1/2 | [37] |
| Human oral cancer (SAS) | In vitro | ↓Nrf-2, ↓p53 | [23] |
| Colorectal cancer (human samples) | In vivo | ↑ERK/SP1/MMP2 | [35] |
3. Mechanisms of HMGB1-RAGE Axis in Cancer Progression
3.1. Beclin-1
3.2. MAPK
3.3. MicroRNA
3.4. MMPs
3.5. NF-κB/Snail
3.6. PI3K/Akt
| Cell Lines/Samples | Study Type | Mechanisms of Action | References |
|---|---|---|---|
| Glioma (C6) | In vitro | ↑ERK1/2, ↑p38, ↑SAPK/JNK | [62] |
| Murine Lewis lung carcinoma | In vitro | ↑ERK1/2, ↑p38, ↑SAPK/JNK | [62] |
| Mouse neuroblastoma (Neuro2a) | In vitro | ↑Bcl-2 | [87] |
| Human neuroblastoma (SH-SY5Y) | In vitro | ↑Bcl-2 | [87] |
| Human colon carcinoma (Colo320 and WiDr) | In vitro | ↑ERK1/2, ↑Rac1, ↑Akt, ↑MMP9 | [13] |
| Human pancreatic carcinoma (PANC-1 and MIA PaCa-2) | In vitro | ↑MMP9 | [71] |
| Human rhabdomyosarcoma (TE671) | In vitro | ↑Cdc42-Rac1-MKK6-p38 | [61] |
| Human oral squamous cell carcinoma (OSCC) | In vitro | ↑VEGF | [83] |
| Human pancreatic adenocarcinoma (Panc 2.03) | In vitro | ↑Beclin-1 | [54] |
| Human oral squamous cell carcinoma (OSCC) | In vitro | ↑VEGF | [84] |
| Human pancreatic cancer (BxPC-3) | In vitro | ↑NF-κB | [74] |
| Human esophageal squamous cell carcinoma (KYSE-150) | In vitro | ↑VEGF-C | [88] |
| Human renal cell carcinoma (CCRCC) | In vitro | ↑ERK1/2 | [56] |
| Human thyroid carcinoma (BC PAP) | In vitro | ↑miR-221/222, ↓PTEN | [66] |
| Human chondrosarcoma (JJ012) | In vitro | ↑PI3K/Akt/c-Jun/AP-1, ↑α5β1 integrin | [81] |
| Human lung cancer (95D) | In vitro | ↑MMP2, ↑MMP9, ↑CDK-2 | [60] |
| Human hepatocarcinoma (HUH 7) | In vitro | ↑NF-κB, ↑p65 | [14] |
| Human hepatocarcinoma (H22) | In vitro | ↑NF-κB, ↑MMP9 | [72] |
| Liver carcinogenesis in mice | In vivo | ↑ERK1/2, ↑Cyclin D1 | [57] |
| Human fibrosarcoma (HT1080) | In vitro | ↑NF-κB | [75] |
| Mouse melanoma (B16-F10) | In vitro | ↑STAT3 | [89] |
| Human non-small cell lung cancer (NSCLC) | In vitro | ↑JNK, ↑NF-κB | [76] |
| Pancreatic tumor (human Panc2.03, human Panc3.27, mouse Panc02) | In vitro | ↑ATP | [90] |
| Human hepatocellular carcinoma (HCC) | In vitro | ↑NF-кB | [77] |
| Murine lung cancer (Lewis cells) | In vitro | ↑PI3K/Akt, ↑ERK1/2, ↑Bcl-2, ↓Bax | [82] |
| Human breast cancer (MCF-7) | In vitro | ↑NF-κB | [78] |
| Human bladder carcinoma (5637, BIU-87, T24, and SV-HUC-1) | In vitro | ↑NF-κB, ↑VEGF | [85] |
| Human thyroid carcinoma (BC PAP) | In vitro | ↑miR-221/222, ↓PTEN | [65] |
| Human nasopharyngeal carcinoma (HONE-1) | In vitro | ↑Bcl-2, ↑p-ERK1/2, ↓caspase-3, ↓Bax | [86] |
| Murine Lung cancer (Lewis cells) | In vitro | ↑ERK1/2 | [91] |
| Human gastric carcinoma (BGC-823, SGC-7901, MKN-28, and MKN-45) | In vitro | ↑ERK1/2 | [58] |
| Human colorectal carcinoma (HCT116 and LoVo) | In vitro | ↑-Snail/NF-κB, ↑MMP7 | [70] |
| Human hepatocellular carcinoma (HCC) | In vitro | ↑MMP2, ↑ERK1/2, ↑p38, ↑SAPK/JNK, ↑MEK1/2, ↑SEK1, ↑c-Jun, ↑c-Myc, ↓p21 | [59] |
| Human colorectal adenocarcinoma (HT-29) | In vitro | ↑MAPK, ↑PI3K | [55] |
| Human breast cancer (MCF-7) | In vitro | ↑MAPK, ↑PI3K | [55] |
| Human adenocarcinomic human alveolar basal epithelial (A549) | In vitro | ↑MAPK, ↑PI3K | [55] |
| Human hypopharyngeal carcinoma (FaDu) | In vitro | ↑Vimentin, ↑Snail | [80] |
| Cervical carcinomas (human specimens and HeLa cells) | In vivo | ↑NF-κB, ↑N-cadherin↓E-cadherin | [79] |
| Human colorectal carcinoma (LoVo) | In vitro | ↑NF-κB | [92] |
| Human prostate cancer (PC-3) | In vitro | ↑NF-κB, ↑MMP1, ↑MMP3, ↑MMP10 | [93] |
4. Mechanisms of S100 Family-RAGE Axis in Cancer Progression
4.1. Angiogenesis
4.2. MAPK
4.3. MMPs
4.4. NF-κB
4.5. p53
4.6. PI3K/Akt/mTOR
4.7. STAT3
| S100 Type | Cell Lines/Samples | Study Type | Mechanisms of Action | References |
|---|---|---|---|---|
| S100A4 | Human osteosarcoma (II-11b) | In vitro | ↑NF-κB | [96] |
| Human melanoma (A375) | In vitro | ↑NF-κB | [127] | |
| Human pancreatic cancer (BxPC-3) | In vitro | ↑NF-κB | [74] | |
| Human pancreatic carcinoma (MiaPACA-2) | In vitro | ↑VEGF | [103] | |
| Human colorectal carcinoma (HCT116, SW620, and DLD-1) | In vitro | ↑ERK | [95] | |
| Human melanoma (B16-F10) | In vitro | ↑NF-κB | [97] | |
| Human colorectal carcinoma (SW480 and LoVo) | In vitro | ↑Akt, ↑mTOR, ↑p70S6K, ↑VEGF, ↓E-cadherin | [101] | |
| Thyroid cancer (human specimens) | In vitro | ↑Cdc42, ↑ERK | [109] | |
| Human melanoma (A375) | In vitro | ↓E-cadherin | [116] | |
| Human melanoma (A375) | In vitro | ↑NF-κB | [128] | |
| S100A6 | Nasopharyngeal carcinoma (human specimens) | In vivo | ↑p38 | [107] |
| S100A7 | Human breast adenocarcinoma (MDA-MB-468) | In vitro | ↑VEGF | [102] |
| Aggressive triple-negative breast cancer (human specimens) | In vivo | ↑ERK, ↑NF-κB, ↑MMP9 | [108] | |
| Human cervical cancer derived (C33A, HeLa, SiHa, and Caski) | In vitro | ↑ERK | [129] | |
| S100A8 | Human prostate cancer (LNCaP and PC-3) | In vitro | ↑NF-κB, ↑p38, ↑ERK1/2 | [105] |
| Esophageal pre-neoplasia in the rat | In vivo | ↑NF-κB | [130] | |
| Colon carcinoma (MC38) | In vitro | ↑NF-κB, ↑ERK1/2, ↑SAPK/JNK | [106] | |
| Oral-esophageal tumor in mice | In vivo | ↑NF-κB | [114] | |
| Human breast cancer (MCF-7 and T47D) | In vitro | ↑NF-κB | [98] | |
| Hepatocellular carcinoma in mice | In vivo | ↑ERK | [131] | |
| Squamous cell carcinoma (human specimens) | In vivo | ↑p38, ↑SAPK/JNK, ↑ERK1/2 | [132] | |
| S100A9 | Human prostate cancer (LNCaP and PC-3) | In vitro | ↑NF-κB, ↑p38, ↑ERK1/2 | [105] |
| Colon carcinoma (MC38) | In vitro | ↑NF-κB, ↑ERK1/2, ↑SAPK/JNK | [106] | |
| Human breast cancer (MCF-7 and T47D) | In vitro | ↑NF-κB | [98] | |
| Hepatocellular carcinoma in mice | In vivo | ↑ERK | [131] | |
| Squamous cell carcinoma (human specimens) | In vivo | ↑p38, ↑SAPK/JNK, ↑ERK1/2 | [132] | |
| Human hepatocellular carcinoma (HepG2, SMMC-7721 and Huh7) | In vitro | ↑p-p38, ↑p-ERK1/2 | [133] | |
| S100A14 | Esophageal squamous cell carcinoma (KYSE180) | In vitro | ↑ERK1/2, ↑NF-κB | [115] |
| S100A16 | Human prostate cancer (DU-145, LNCaP, and PC-3) | In vitro | ↑Akt, ↑ERK, ↓p21, ↓p27 | [121] |
| S100B | Human melanoma (WM115) | In vitro | ↓p53 | [119] |
| Human large cell lung carcinoma (H1299) | In vitro | ↓p53 | [117] | |
| Human breast cancer (MCF-7) | In vitro | ↓p53 | [117] | |
| Human colorectal carcinoma (SW480) | In vitro | ↑ERK1/2 | [134] | |
| Human malignant melanoma (C8146A) | In vitro | ↓p53 | [118] | |
| Human neuroblastoma (SH-SY5Y) | In vitro | ↑PI3K/Akt, ↑NF-κB | [113] | |
| Human malignant melanoma (C8146A) | In vitro | ↓p53 | [135] | |
| Murine glioma (GL261) | In vitro | ↑STAT3 | [125] | |
| Ovarian cancer stem-like cell | In vitro | ↓p53 | [120] | |
| Glioma (C6) | In vitro | ↑Akt1, ↑STAT3 | [124] | |
| S100P | Human pancreatic adenocarcinoma (BxPC-3 and MPanc-96) | In vitro | ↑NF-κB | [136] |
| Human colon cancer (SW480) | In vitro | ↑NF-κB, ↑ERK1/2 | [104] | |
| Human pancreatic cancer (BxPC-3) | In vitro | ↑NF-κB | [74] | |
| Human colorectal carcinoma (LS174T and SW480) | In vitro | ↑miR-155 | [137] | |
| Human pancreatic carcinoma (BxPC3) | In vitro | ↑MMP9 | [111] | |
| Human oral squamous cell carcinoma (OSCC) | In vitro | ↑NF-κB | [138] | |
| Human colorectal carcinoma (SW480 and LS174T) | In vitro | ↑c-Fos, ↑AP-1, ↑miR-21 | [139] | |
| Human nasopharyngeal carcinoma (C666-1) | In vitro | ↑MMP2, ↑MMP9 | [112] |
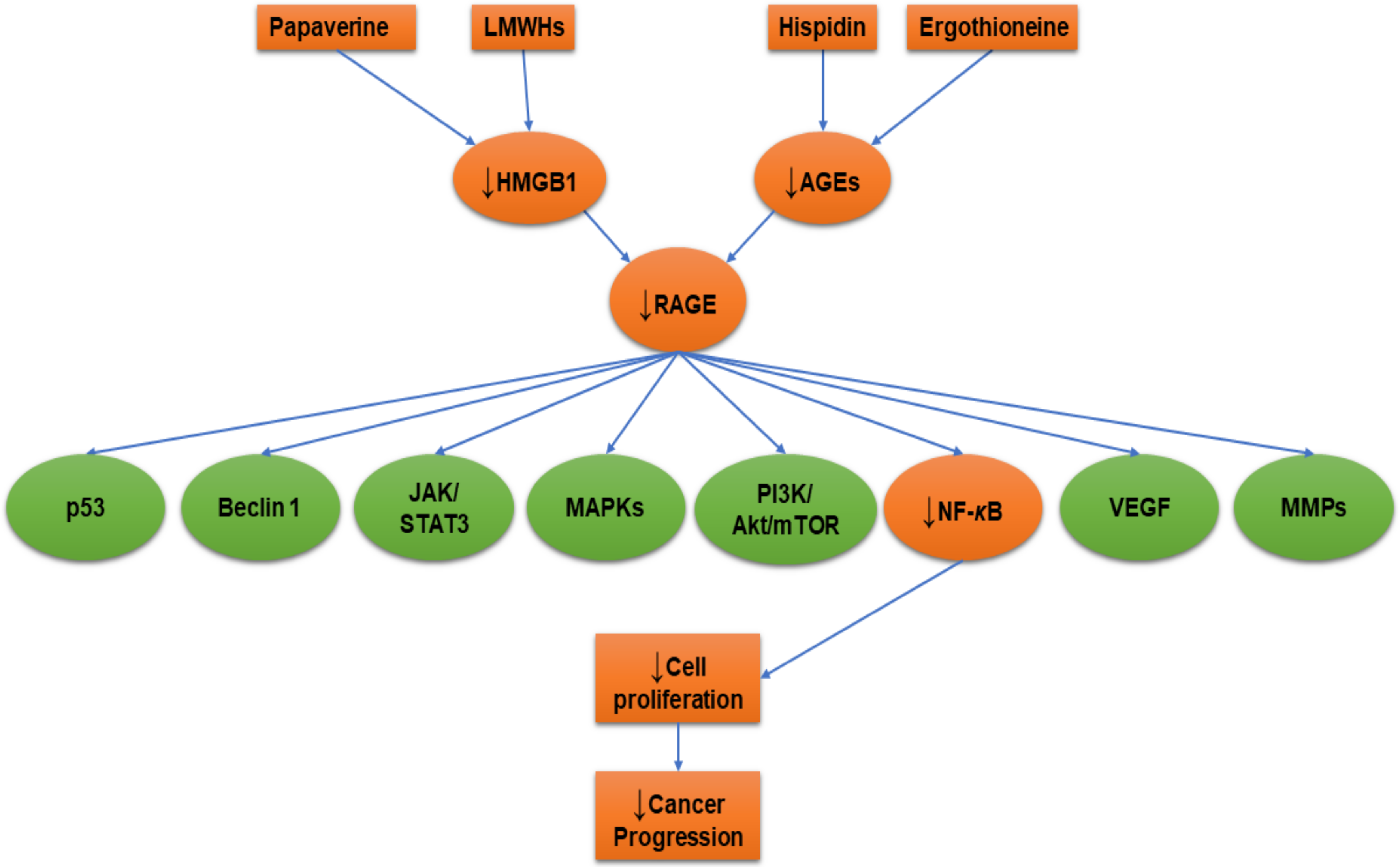
5. RAGE-Inhibitors
5.1. Duloxetine
5.2. Ethyl Pyruvate
5.3. Hispidin
5.4. Heparin
5.5. Papaverine
| Drug | Cell Line/Samples | Study Type | Mechanisms of Action | References |
|---|---|---|---|---|
| Chondroitin sulfate and heparan sulfate | Mice | In vivo | ↓Lung metastasis | [152] |
| Duloxetine | Mouse glioma cells (GL261)/mice | In vitro/In vivo | ↓S100B | [144] |
| Ergothioneine | Rat pheochromocytoma (PC12) | In vitro | ↓AGEs, ↓RAGE, ↓NF-κB | [44] |
| Ethyl pyruvate | Human malignant mesothelioma (MM) | In vitro | ↓HMBG1, ↓RAGE, ↓NF-κB | [145] |
| Non-small cell lung cancer (A549, H520, and PC-9) | In vitro | ↓HMGB1, ↓RAGE, ↓NF-κB, ↓STAT3 | [147] | |
| Hispidin | Rat pheochromocytoma (PC12) | In vitro | ↓AGEs, ↓RAGE, ↓NF-κB | [44] |
| Heparin (low molecular weight) | Human fibrosarcoma (HT1080) | In vitro | ↓NF-κB | [75] |
| Papaverine | Human fibrosarcoma (HT1080) | In vitro | ↓HMBG1, ↓RAGE, ↓NF-κB | [11] |
| Human glioblastoma (U87MG and T98G) | ↓HMBG1, ↓RAGE | [153] |
6. Conclusions
- ▪
- Comparative studies of RAGE-ligands.
- ▪
- The role of RAGE-ligands in cancer progression in primary cell culture of surgically removed tumor masses or cancer biopsies.
- ▪
- The role of RAGE-ligands in cancer progression using cancer stem cells.
- ▪
- The role of AGEs in colorectal cancer with therapeutic trials.
- ▪
- Studies of the effect of RAGE-ligands’ pathway signaling on intrinsic pathway components such as cytochrome c, apoptotic protease activating factor 1 (Apaf-1), caspase-9, and caspase-3.
- ▪
- Studies of the effect of RAGE-ligands’ pathway signaling on extrinsic pathway components such as tumor necrosis factor receptor-associated death domain (TRADD), Fas-associated death domain (FADD), caspase-8, and caspase-10.
- ▪
- Studies of the effect of RAGE-ligands’ pathway signaling on Bcl-2 family, either the pro-apoptotic (BAX, BID, BAK, or BAD) or anti-apoptotic (Bcl-Xl and Bcl-2).
- ▪
- Studies of the effect of RAGE-ligands’ pathway signaling on molecules that induce cell survival and metastasis including E-cadherin, hypoxia-inducible factor 1-alpha (HIF-1α), PTEN, and MDM2.
- ▪
- Studies of the effect of RAGE-ligands’ pathway signaling on cyclin-dependent kinases (CDK-1, 2, 4, or 6) and regulatory cyclin subunits (cyclin A, B, Ds, or E).
- ▪
- Studies of the effect of RAGE-ligands’ pathway signaling on molecules that facilitate cell survival and metastasis such as β-catenin, epidermal growth factor receptor (EGFR), VEGF, and vimentin.
- ▪
- Discovery of new drugs that downregulate RAGE and its ligands to control cancer progression.
- ▪
- The role of RAGE-ligands in cancer senescence and senotherapies.
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Akt | protein kinase B |
| AP-1 | activator protein 1 |
| Apaf-1 | apoptotic protease activating factor 1 |
| ATP | adenosine triphosphate |
| Bad | Bcl-2-associated death promoter, |
| Bak | Bcl-2 homologous antagonist/killer |
| Bax | Bcl-2-associated X protein |
| Bcl-2; | B-cell lymphoma 2 |
| Bcl-XL | B-cell lymphoma-extra-large |
| Cdc42 | cell division control protein 42 homolog |
| CDK-1 | cyclin-dependent kinase-1 |
| CDK-2 | cyclin-dependent kinase-2 |
| CDK-4 | cyclin-dependent kinase-4 |
| CDK-6 | cyclin-dependent kinase-6 |
| ChREBP | carbohydrate responsive element binding protein |
| EGFR | epidermal growth factor receptor |
| ERK | extracellular signal-regulated kinase |
| FADD | Fas-associated death domain |
| HIF-1α | hypoxia-inducible factor 1-alpha |
| HMGB1 | high-mobility group box1 |
| JAK | Janus kinase |
| JNK | c-Jun N-terminal kinase |
| MAPK | mitogen-activated protein kinase |
| MDM2 | mouse double minute 2 homolog |
| MKK6 | mitogen-activated protein kinase kinase 6 |
| MMPs | matrix metalloproteinases |
| mTOR | mammalian target of rapamycin, |
| Nrf-2 | Nuclear factor (erythroid-derived 2)-like 2 |
| NF-κB | nuclear factor kappa B |
| p70S6K | ribosomal protein S6 kinase B1 |
| PI3K | phosphatidylinositide 3-kinase |
| PTEN | phosphatase and tensin homolog |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 |
| RAGE | receptor of advanced glycation end product |
| ROS | reactive oxygen species |
| SAPK | stress-activated protein kinases |
| STAT3 | signal transducer and activator of transcription 3 |
| TRADD | tumor necrosis factor receptor-associated death domain |
| VEGF | vascular endothelial growth factor |
| VEGF-C | vascular endothelial growth factor C. |
References
- Iwamura, M.; Yamamoto, Y.; Kitayama, Y.; Higuchi, K.; Fujimura, T.; Hase, T.; Yamamoto, H. Epidermal expression of receptor for advanced glycation end products (RAGE) is related to inflammation and apoptosis in human skin. Exp. Dermatol. 2016, 25, 235–237. [Google Scholar] [CrossRef] [PubMed]
- López-Díez, R.; Rastrojo, A.; Villate, O.; Aguado, B. Complex Tissue-Specific Patterns and Distribution of Multiple RAGE Splice Variants in Different Mammals. Genome Biol. Evol. 2013, 5, 2420–2435. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.I.; Carter, A.M.; Harja, E.; Kalea, A.Z.; Arriero, M.; Yang, H.; Grant, P.J.; Schmidt, A.M. Identification, classification, and expression of RAGE gene splice variants. FASEB J. 2008, 22, 1572–1580. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Keller, J.N. Evaluation of rage isoforms, ligands, and signaling in the brain. Biochim. Biophys. Acta Mol. Cell Res. 2005, 1746, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Oury, T.D.; Li, Y.; Liu, S.; Zhang, Z.; Xu, Q.; Xie, F.; Wang, J.; Ping, S.; Li, C.; Wang, Z.; et al. RAGE Mediates Accelerated Diabetic Vein Graft Atherosclerosis Induced by Combined Mechanical Stress and AGEs via Synergistic ERK Activation. PLoS ONE 2012, 7, e35016. [Google Scholar] [CrossRef]
- He, M.; Kubo, H.; Morimoto, K.; Fujino, N.; Suzuki, T.; Takahasi, T.; Yamada, M.; Yamaya, M.; Maekawa, T.; Yamamoto, Y.; et al. Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells. EMBO Rep. 2011, 12, 358–364. [Google Scholar] [CrossRef]
- Win, M.T.T.; Yamamoto, Y.; Munesue, S.; Saito, H.; Han, D.; Motoyoshi, S.; Kamal, T.; Ohara, T.; Watanabe, T.; Yamamoto, H.; et al. Regulation of RAGE for attenuating progression of diabetic vascular complications. Exp. Diabetes Res. 2012, 2012, 894605. [Google Scholar] [CrossRef]
- Yan, S.D.; Chen, X.; Fu, J.; Chen, M.; Zhu, H.; Roher, A.; Slattery, T.; Zhao, L.; Nagashima, M.; Morser, J.; et al. RAGE and amyloid-β peptide neurotoxicity in Alzheimer’s disease. Nature 1996, 382, 685–691. [Google Scholar] [CrossRef]
- Huttunen, H.J.; Fages, C.; Rauvala, H. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-kappaB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 1999, 274, 19919–19924. [Google Scholar] [CrossRef]
- Lander, H.M.; Tauras, J.M.; Ogiste, J.S.; Hori, O.; Moss, R.A.; Schmidt, A.M. Activation of the receptor for advanced glycation end products triggers a p21(ras)-dependent mitogen-activated protein kinase pathway regulated by oxidant stress. J. Biol. Chem. 1997, 272, 17810–17814. [Google Scholar] [CrossRef]
- El-Far, A.; Munesue, S.; Harashima, A.; Sato, A.; Shindo, M.; Nakajima, S.; Inada, M.; Tanaka, M.; Takeuchi, A.; Tsuchiya, H.; et al. In vitro anticancer effects of a RAGE inhibitor discovered using a structure-based drug design system. Oncol. Lett. 2018, 15, 4627–4634. [Google Scholar] [CrossRef] [PubMed]
- Flohr, A.M.; Rogalla, P.; Meiboom, M.; Borrmann, L.; Krohn, M.; Thode-Halle, B.; Bullerdiek, J. Variation of HMGB1 expression in breast cancer. Anticancer Res. 2001, 21, 3881–3885. [Google Scholar] [PubMed]
- Kuniyasu, H.; Chihara, Y.; Kondo, H. Differential effects between amphoterin and advanced glycation end products on colon cancer cells. Int. J. Cancer 2003, 104. [Google Scholar] [CrossRef] [PubMed]
- Yaser, A.-M.; Huang, Y.; Zhou, R.-R.; Hu, G.-S.; Xiao, M.-F.; Huang, Z.-B.; Duan, C.-J.; Tian, W.; Tang, D.-L.; Fan, X.-G. The Role of Receptor for Advanced Glycation End Products (RAGE) in the Proliferation of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2012, 13, 5982–5997. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.; Jurgensmeier, J.; Matsuyama, S. Bcl-2 family proteins and mitochondria. Biochim. Biophys. Acta Bioenerg. 1998, 1366, 127–137. [Google Scholar] [CrossRef]
- El-Far, A.H.; Badria, F.A.; Shaheen, H.M. Possible Anticancer Mechanisms of Some Costus speciosus Active Ingredients Concerning Drug Discovery. Curr. Drug Discov. Technol. 2016, 13, 123–143. [Google Scholar] [CrossRef]
- Evan, G.I.; Vousden, K.H. Proliferation, cell cycle and apoptosis in cancer. Nature 2001, 411, 342–348. [Google Scholar] [CrossRef]
- Kang, R.; Tang, D.; Lotze, M.T.; Zeh, H.J. RAGE regulates autophagy and apoptosis following oxidative injury. Autophagy 2011, 7, 442–444. [Google Scholar] [CrossRef]
- Ahmed, N. Advanced glycation endproducts—Role in pathology of diabetic complications. Diabetes Res. Clin. Pract. 2005, 67, 3–21. [Google Scholar] [CrossRef]
- Peyroux, J.; Sternberg, M. Advanced glycation endproducts (AGEs): Pharmacological inhibition in diabetes. Pathol. Biol. 2006, 54, 405–419. [Google Scholar] [CrossRef]
- Takino, J.; Yamagishi, S.; Takeuchi, M. Glycer-AGEs-RAGE signaling enhances the angiogenic potential of hepatocellular carcinoma by upregulating VEGF expression. World J. Gastroenterol. 2012, 18, 1781–1788. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.-M.; He, M.-Y.; Liu, Y.-W.; Lu, Y.-J.; Hong, Y.-Q.; Luo, H.-H.; Ren, Z.-L.; Zhao, S.-C.; Jiang, Y. AGE/RAGE/Akt pathway contributes to prostate cancer cell proliferation by promoting Rb phosphorylation and degradation. Am. J. Cancer Res. 2015, 5, 1741–1750. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.-Y.; Ko, H.-A.; Shieh, T.-M.; Chi, T.-C.; Chen, H.-I.; Chen, Y.-T.; Yu, Y.-H.; Yang, S.-H.; Chang, S.-S. Advanced glycation end products influence oral cancer cell survival via Bcl-xl and Nrf-2 regulation In Vitro. Oncol. Lett. 2017, 13, 3328–3334. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Matsui, T.; Takeuchi, M.; Yamagishi, S. Metformin Inhibits Advanced Glycation End Products (AGEs)-induced Growth and VEGF Expression in MCF-7 Breast Cancer Cells by Suppressing AGEs Receptor Expression via AMP-activated Protein Kinase. Horm. Metab. Res. 2012, 45, 387–390. [Google Scholar] [CrossRef]
- Sharaf, H.; Matou-Nasri, S.; Wang, Q.; Rabhan, Z.; Al-Eidi, H.; Al Abdulrahman, A.; Ahmed, N. Advanced glycation endproducts increase proliferation, migration and invasion of the breast cancer cell line MDA-MB-231. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 429–441. [Google Scholar] [CrossRef]
- Wang, X.; Yu, S.; Wang, C.-Y.; Wang, Y.; Liu, H.-X.; Cui, Y.; Zhang, L.-D. Advanced glycation end products induce oxidative stress and mitochondrial dysfunction in SH-SY5Y cells. Vitr. Cell. Dev. Biol. Anim. 2014, 51, 204–209. [Google Scholar] [CrossRef]
- Zill, H.; Günther, R.; Erbersdobler, H.F.; Fölsch, U.R.; Faist, V. RAGE Expression and AGE-Induced MAP Kinase Activation in Caco-2 Cells. Biochem. Biophys. Res. Commun. 2001, 288, 1108–1111. [Google Scholar] [CrossRef]
- Chen, H.; Wu, L.; Li, Y.; Meng, J.; Lin, N.; Yang, D.; Zhu, Y.; Li, X.; Li, M.; Xu, Y.; et al. Advanced glycation end products increase carbohydrate responsive element binding protein expression and promote cancer cell proliferation. Mol. Cell. Endocrinol. 2014, 395, 69–78. [Google Scholar] [CrossRef]
- Iizuka, K.; Horikawa, Y. ChREBP: A Glucose-activated Transcription Factor Involved in the Development of Metabolic Syndrome. Endocr. J. 2008, 55, 617–624. [Google Scholar] [CrossRef]
- Tong, X.; Zhao, F.; Mancuso, A.; Gruber, J.J.; Thompson, C.B. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc. Natl. Acad. Sci. USA 2009, 106, 21660–21665. [Google Scholar] [CrossRef]
- Aaronson, D.S. A Road Map for Those Who Don’t Know JAK-STAT. Science 2002, 296, 1653–1655. [Google Scholar] [CrossRef]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.P.; Danson, S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, H.K.; Yoon, J.S.; Kim, S.J.; Kim, E.S.; Ahn, K.S.; Kim, D.-S.; Yoon, S.S.; Kim, B.K.; Lee, Y.Y. Advanced glycation end product (AGE)-induced proliferation of HEL cells via receptor for AGE-related signal pathways. Int. J. Oncol. 2008, 33, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Mo, F.; Chang, B.; Zhang, Q.; Ran, H.; Yang, S.; Zhu, Z.; Hu, L.; Su, Q. Glucose-derived AGEs enhance human gastric cancer metastasis through RAGE/ERK/Sp1/MMP2 cascade. Oncotarget 2017, 8, 104216–104226. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Wu, H.; Ran, H.; Kong, X.; Hu, L.; Wang, X.; Su, Q. Glucose-derived AGEs promote migration and invasion of colorectal cancer by up-regulating Sp1 expression. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.I.; Kalea, A.Z.; del Mar Arriero, M.; Harja, E.; Boulanger, E.; D’Agati, V.; Schmidt, A.M. Interaction of the RAGE Cytoplasmic Domain with Diaphanous-1 Is Required for Ligand-stimulated Cellular Migration through Activation of Rac1 and Cdc42. J. Biol. Chem. 2008, 283, 34457–34468. [Google Scholar] [CrossRef]
- Matou-Nasri, S.; Sharaf, H.; Wang, Q.; Almobadel, N.; Rabhan, Z.; Al-Eidi, H.; Yahya, W.B.; Trivilegio, T.; Ali, R.; Al-Shanti, N.; et al. Biological impact of advanced glycation endproducts on estrogen receptor-positive MCF-7 breast cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2808–2820. [Google Scholar] [CrossRef]
- Mook, O.R.F.; Frederiks, W.M.; Van Noorden, C.J.F. The role of gelatinases in colorectal cancer progression and metastasis. Biochim. Biophys. Acta Rev. Cancer 2004, 1705, 69–89. [Google Scholar] [CrossRef]
- Ko, S.S.-Y.; Ko, H.-A.; Shieh, T.-M.; Chang, W.-C.; Chen, H.-I.; Chang, S.S.-S.; Lin, I.-H.; Ahmad, S.K.; Moinuddin, D.; Shahab, U.; et al. Cell Migration Is Regulated by AGE-RAGE Interaction in Human Oral Cancer Cells In Vitro. PLoS ONE 2014, 9, e110542. [Google Scholar] [CrossRef]
- Sajadimajd, S.; Khazaei, M. Oxidative Stress and Cancer: The Role of Nrf2. Curr. Cancer Drug Targets 2018, 18, 538–557. [Google Scholar] [CrossRef]
- Lee, Y.-M.; Auh, Q.-S.; Lee, D.-W.; Kim, J.-Y.; Jung, H.-J.; Lee, S.-H.; Kim, E.-C. Involvement of Nrf2-Mediated Upregulation of Heme Oxygenase-1 in Mollugin-Induced Growth Inhibition and Apoptosis in Human Oral Cancer Cells. Biomed Res. Int. 2013, 2013, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Korwar, A.M.; Bhonsle, H.S.; Chougale, A.D.; Kote, S.S.; Gawai, K.R.; Ghole, V.S.; Koppikar, C.B.; Kulkarni, M.J. Analysis of AGE modified proteins and RAGE expression in HER2/neu negative invasive ductal carcinoma. Biochem. Biophys. Res. Commun. 2012, 419, 490–494. [Google Scholar] [CrossRef] [PubMed]
- Munesue, S.; Yamamoto, Y.; Urushihara, R.; Inomata, K.; Saito, H.; Motoyoshi, S.; Watanabe, T.; Yonekura, H.; Yamamoto, H. Low-molecular weight fractions of Japanese soy sauce act as a RAGE antagonist via inhibition of RAGE trafficking to lipid rafts. Food Funct. 2013, 4, 1835. [Google Scholar] [CrossRef] [PubMed]
- Song, T.-Y.; Yang, N.-C.; Chen, C.-L.; Thi, T.L.V. Protective Effects and Possible Mechanisms of Ergothioneine and Hispidin against Methylglyoxal-Induced Injuries in Rat Pheochromocytoma Cells. Oxid. Med. Cell. Longev. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.S.; Huang, Y. Death Receptor Activation Complexes: It Takes Two to Activate TNF Receptor 1. Cell Cycle 2014, 2, 549–551. [Google Scholar] [CrossRef]
- Carmeliet, P. VEGF as a Key Mediator of Angiogenesis in Cancer. Oncology 2005, 69, 4–10. [Google Scholar] [CrossRef]
- Choi, Y.R.; Kim, H.; Kang, H.J.; Kim, N.-G.; Kim, J.J.; Park, K.-S.; Paik, Y.-K.; Kim, H.O.; Kim, H. Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation. Cancer Res. 2003, 63, 2188–2193. [Google Scholar]
- Volp, K.; Völp, K.; Brezniceanu, M.-L.; Bösser, S.; Brabletz, T.; Kirchner, T.; Göttel, D.; Joos, S.; Zörnig, M. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut 2006, 55, 234–242. [Google Scholar] [CrossRef]
- Yan, W.; Chang, Y.; Liang, X.; Cardinal, J.S.; Huang, H.; Thorne, S.H.; Monga, S.P.S.; Geller, D.A.; Lotze, M.T.; Tsung, A. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology 2012, 55, 1863–1875. [Google Scholar] [CrossRef]
- Zhang, Q.-B.; Jia, Q.; Wang, H.; Hu, C.-X.; Sun, D.; Jiang, R.-D.; Zhang, Z.-L.; El-Serag, H.; Rudolph, K.; Rampone, B.; et al. High-mobility group protein box1 expression correlates with peritumoral macrophage infiltration and unfavorable prognosis in patients with hepatocellular carcinoma and cirrhosis. BMC Cancer 2016, 16, 880. [Google Scholar] [CrossRef]
- Smolarczyk, R.; Cichoń, T.; Jarosz, M.; Szala, S. HMGB1—Its role in tumor progression and anticancer therapy. Postepy Hig. Med. Dosw. 2012, 66, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.A.; Shaw, L.M. An autophagy-independent function of Beclin 1 in cancer. Mol. Cell. Oncol. 2016, 3, e1030539. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.A.; Janusis, J.; Leonard, D.; Bellvé, K.D.; Fogarty, K.E.; Baehrecke, E.H.; Corvera, S.; Shaw, L.M. Beclin 1 regulates growth factor receptor signaling in breast cancer. Oncogene 2015, 34, 5352–5362. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Cheh, C.-W.; Livesey, K.M.; Liang, X.; Schapiro, N.E.; Benschop, R.; Sparvero, L.J.; Amoscato, A.A.; Tracey, K.J.; et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene 2010, 29, 5299–5310. [Google Scholar] [CrossRef]
- Sharma, S.; Evans, A.; Hemers, E. Mesenchymal-epithelial signalling in tumour microenvironment: Role of high-mobility group Box 1. Cell Tissue Res. 2016, 365, 357–366. [Google Scholar] [CrossRef]
- Lin, L.; Zhong, K.; Sun, Z.; Wu, G.; Ding, G. Receptor for advanced glycation end products (RAGE) partially mediates HMGB1-ERKs activation in clear cell renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2012, 138, 11–22. [Google Scholar] [CrossRef]
- Pusterla, T.; Nèmeth, J.; Stein, I.; Wiechert, L.; Knigin, D.; Marhenke, S.; Longerich, T.; Kumar, V.; Arnold, B.; Vogel, A.; et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology 2013, 58, 363–373. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Wu, L.Q.; Zhang, T.; Han, Y.F.; Lin, X. Autophagy-mediated HMGB1 release promotes gastric cancer cell survival via RAGE activation of extracellular signal-regulated kinases 1/2. Oncol. Rep. 2015, 33, 1630–1638. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, C.; Liu, Y.; Jiang, Y. HMGB1 promotes HCC progression partly by downregulating p21 via ERK/c-Myc pathway and upregulating MMP-2. Tumour Biol. 2016, 37, 4399–4408. [Google Scholar] [CrossRef]
- Wang, C.; Fei, G.; Liu, Z.; Li, Q.; Xu, Z.; Ren, T. HMGB1 was a pivotal synergistic effecor for CpG oligonucleotide to enhance the progression of human lung cancer cells. Cancer Biol. Ther. 2012, 13, 727–736. [Google Scholar] [CrossRef]
- Riuzzi, F.; Sorci, G.; Donato, R. RAGE expression in rhabdomyosarcoma cells results in myogenic differentiation and reduced proliferation, migration, invasiveness, and tumor growth. Am. J. Pathol. 2007, 171, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, A.; Blood, D.C.; del Toro, G.; Canet, A.; Lee, D.C.; Qu, W.; Tanji, N.; Lu, Y.; Lalla, E.; Fu, C.; et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 2000, 405, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, K.; Sivasankar, V. MicroRNAs—Biology and clinical applications. J. Oral Maxillofac. Pathol. 2014, 18, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Mardente, S.; Mari, E.; Massimi, I.; Fico, F.; Faggioni, A.; Pulcinelli, F.; Antonaci, A.; Zicari, A. HMGB1-Induced Cross Talk between PTEN and miRs 221/222 in Thyroid Cancer. Biomed Res. Int. 2015, 2015, 1–7. [Google Scholar] [CrossRef]
- Mardente, S.; Mari, E.; Consorti, F.; Di Gioia, C.; Negri, R.; Etna, M.; Zicari, A.; Antonaci, A. HMGB1 induces the overexpression of miR-222 and miR-221 and increases growth and motility in papillary thyroid cancer cells. Oncol. Rep. 2012, 28, 2285–2289. [Google Scholar] [CrossRef]
- Jikuzono, T.; Kawamoto, M.; Yoshitake, H.; Kikuchi, K.; Akasu, H.; Ishikawa, H.; Hirokawa, M.; Miyauchi, A.; Tsuchiya, S.; Shimizu, K.; et al. The miR-221/222 cluster, miR-10b and miR-92a are highly upregulated in metastatic minimally invasive follicular thyroid carcinoma. Int. J. Oncol. 2013, 42, 1858–1868. [Google Scholar] [CrossRef]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef]
- Winer, A.; Adams, S.; Mignatti, P. Matrix metalloproteinase inhibitors in cancer therapy: Turning past failures into future successes. Mol. Cancer Ther. 2018, 17, 1147–1155. [Google Scholar] [CrossRef]
- Zhu, L.; Li, X.; Chen, Y.; Fang, J.; Ge, Z. High-mobility group Box 1: A novel inducer of the epithelial-mesenchymal transition in colorectal carcinoma. Cancer Lett. 2015, 357, 527–534. [Google Scholar] [CrossRef]
- Takada, M.; Hirata, K.; Ajiki, T.; Suzuki, Y.; Kuroda, Y. Expression of receptor for advanced glycation end products (RAGE) and MMP-9 in human pancreatic cancer cells. Hepatogastroenterology 2004, 51, 928–930. [Google Scholar] [PubMed]
- Gong, W.; Wang, Z.-Y.; Chen, G.-X.; Liu, Y.-Q.; Gu, X.-Y.; Liu, W.-W. Invasion potential of H22 hepatocarcinoma cells is increased by HMGB1-induced tumor NF-κB signaling via initiation of HSP70. Oncol. Rep. 2013, 30, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Saarialho-Kere, U.K.; Chang, E.S.; Welgus, H.G.; Parks, W.C. Distinct localization of collagenase and tissue inhibitor of metalloproteinases expression in wound healing associated with ulcerative pyogenic granuloma. J. Clin. Invest. 1992, 90, 1952–1957. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, T.; Ramachandran, V.; Gomez, S.B.; Schmidt, A.M.; Logsdon, C.D. S100P-derived RAGE antagonistic peptide reduces tumor growth and metastasis. Clin. Cancer Res. 2012, 18, 4356–4364. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, A.; Yamamoto, Y.; Munesue, S.; Harashima, A.; Watanabe, T.; Yonekura, H.; Yamamoto, H.; Tsuchiya, H. Low molecular weight heparin suppresses receptor for advanced glycation end products-mediated expression of malignant phenotype in human fibrosarcoma cells. Cancer Sci. 2013, 104, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Mi, Y.; Yang, H.; Hu, A.; Zhang, Q.; Shang, C. The activation of HMGB1 as a progression factor on inflammation response in normal human bronchial epithelial cells through RAGE/JNK/NF-κB pathway. Mol. Cell. Biochem. 2013, 380, 249–257. [Google Scholar] [CrossRef]
- Chen, R.; Yi, P.; Zhou, R.; Xiao, M.; Huang, Z.; Tang, D.; Huang, Y.; Fan, X. The role of HMGB1-RAGE axis in migration and invasion of hepatocellular carcinoma cell lines. Mol. Cell. Biochem. 2014, 390, 271–280. [Google Scholar] [CrossRef]
- Dhumale, S.S.; Waghela, B.N.; Pathak, C. Quercetin protects necrotic insult and promotes apoptosis by attenuating the expression of RAGE and its ligand HMGB1 in human breast adenocarcinoma cells. IUBMB Life 2015, 67, 361–373. [Google Scholar] [CrossRef]
- Pang, X.; Zhang, Y.; Zhang, S. High-mobility group box 1 is overexpressed in cervical carcinoma and promotes cell invasion and migration in vitro. Oncol. Rep. 2017, 37, 831–840. [Google Scholar] [CrossRef]
- Li, Y.; Wang, P.; Zhao, J.; Li, H.; Liu, D.; Zhu, W. HMGB1 attenuates TGF-β-induced epithelial-mesenchymal transition of FaDu hypopharyngeal carcinoma cells through regulation of RAGE expression. Mol. Cell. Biochem. 2017, 431, 1–10. [Google Scholar] [CrossRef]
- Tang, C.-H.; Keng, Y.-T.; Liu, J.-F. HMGB-1 induces cell motility and α5β1 integrin expression in human chondrosarcoma cells. Cancer Lett. 2012, 322, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhu, H.; Wang, T.; Sun, Y.; Ni, P.; Liu, Y.; Tian, S.; Amoah Barnie, P.; Shen, H.; Xu, W.; et al. Exogenous High-Mobility Group Box 1 Inhibits Apoptosis and Promotes the Proliferation of Lewis Cells via RAGE/TLR4-Dependent Signal Pathways. Scand. J. Immunol. 2014, 79, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Sasahira, T.; Kirita, T.; Oue, N.; Bhawal, U.K.; Yamamoto, K.; Fujii, K.; Ohmori, H.; Luo, Y.; Yasui, W.; Bosserhoff, A.K.; et al. High mobility group box-1-inducible melanoma inhibitory activity is associated with nodal metastasis and lymphangiogenesis in oral squamous cell carcinoma. Cancer Sci. 2008, 99, 1806–1812. [Google Scholar] [CrossRef] [PubMed]
- Sasahira, T.; Yamamoto, K.; Kurihara, M.; Bhawal, U.K.; Chihara, Y.; Kirita, T.; Kuniyasu, H. The roles of HMGB1 related angiogenesis and lymphangiogenesis in oral cancer. Oncol. Rev. 2010, 5, 49–55. [Google Scholar] [CrossRef]
- Huang, Z.; Zhong, Z.; Zhang, L.; Wang, X.; Xu, R.; Zhu, L.; Wang, Z.; Hu, S.; Zhao, X.; Babjuk, M.; et al. Down-regulation of HMGB1 expression by shRNA constructs inhibits the bioactivity of urothelial carcinoma cell lines via the NF-κB pathway. Sci. Rep. 2015, 5, 12807. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Hu, M.; Wu, T.; Chen, Z.; Zhang, C.; Huang, S.; Zhou, X. Effects of high-mobility group box 1 knockdown on proliferation, migration and invasion of the HONE-1 human nasopharyngeal carcinoma cell line. Mol. Med. Rep. 2015, 12, 7531–7537. [Google Scholar] [CrossRef][Green Version]
- Sajithlal, G.; Huttunen, H.; Rauvala, H.; Munch, G. Receptor for Advanced Glycation End Products Plays a More Important Role in Cellular Survival than in Neurite Outgrowth during Retinoic Acid-induced Differentiation of Neuroblastoma Cells. J. Biol. Chem. 2002, 277, 6888–6897. [Google Scholar] [CrossRef]
- Chen, C.; Tang, P.; Yu, Z. [Effect of HMGB1 on the VEGF-C expression and proliferation of esophageal squamous cancer cells]. Zhonghua Zhong Liu Za Zhi 2012, 34, 566–570. [Google Scholar]
- Tang, Q.; Li, J.; Zhu, H.; Li, P.; Zou, Z.; Xiao, Y. Hmgb1-IL-23-IL-17-IL-6-Stat3 Axis Promotes Tumor Growth in Murine Models of Melanoma. Mediators Inflamm. 2013, 2013, 1–13. [Google Scholar] [CrossRef]
- Kang, R.; Tang, D.; Schapiro, N.E.; Loux, T.; Livesey, K.M.; Billiar, T.R.; Wang, H.; Van Houten, B.; Lotze, M.T.; Zeh, H.J. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 2013, 33, 567–577. [Google Scholar] [CrossRef]
- Su, Z.; Wang, T.; Zhu, H.; Zhang, P.; Han, R.; Liu, Y.; Ni, P.; Shen, H.; Xu, W.; Xu, H. HMGB1 modulates Lewis cell autophagy and promotes cell survival via RAGE-HMGB1-Erk1/2 positive feedback during nutrient depletion. Immunobiology 2015, 220, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Geng, Y.; Deng, Q.; Li, R.; Shao, X.; Zhang, Z.; Xu, W.; Wu, Y.; Ma, Q. Translationally controlled tumor protein affects colorectal cancer metastasis through the high mobility group box 1-dependent pathway. Int. J. Oncol. 2018, 53, 1149–1481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shao, S.; Han, D.; Xu, Y.; Jiao, D.; Wu, J.; Yang, F.; Ge, Y.; Shi, S.; Li, Y.; et al. High mobility group box 1 promotes the epithelial-to-mesenchymal transition in prostate cancer PC3 cells via the RAGE/NF-κB signaling pathway. Int. J. Oncol. 2018, 53, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef]
- Dahlmann, M.; Okhrimenko, A.; Marcinkowski, P.; Osterland, M.; Herrmann, P.; Smith, J.; Heizmann, C.W.; Schlag, P.M.; Stein, U. RAGE mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognostic biomarker for human colorectal cancer metastasis. Oncotarget 2014, 5, 3220–3233. [Google Scholar] [CrossRef]
- Grotterød, I.; Mælandsmo, G.M.; Boye, K. Signal transduction mechanisms involved in S100A4-induced activation of the transcription factor NF-κB. BMC Cancer 2010, 10, 241. [Google Scholar] [CrossRef]
- Haase-Kohn, C.; Wolf, S.; Herwig, N.; Mosch, B.; Pietzsch, J. Metastatic potential of B16-F10 melanoma cells is enhanced by extracellular S100A4 derived from RAW264.7 macrophages. Biochem. Biophys. Res. Commun. 2014, 446, 143–148. [Google Scholar] [CrossRef]
- Yin, C.; Li, H.; Zhang, B.; Liu, Y.; Lu, G.; Lu, S.; Sun, L.; Qi, Y.; Li, X.; Chen, W. RAGE-binding S100A8/A9 promotes the migration and invasion of human breast cancer cells through actin polymerization and epithelial-mesenchymal transition. Breast Cancer Res. Treat. 2013, 142, 297–309. [Google Scholar] [CrossRef]
- Ramjiawan, R.R.; Griffioen, A.W.; Duda, D.G. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis 2017, 20, 185–204. [Google Scholar] [CrossRef]
- Ferrara, N. Vascular Endothelial Growth Factor as a Target for Anticancer Therapy. Oncologist 2004, 9, 2–10. [Google Scholar] [CrossRef]
- Wang, H.; Duan, L.; Zou, Z.; Li, H.; Yuan, S.; Chen, X.; Zhang, Y.; Li, X.; Sun, H.; Zha, H.; et al. Activation of the PI3K/Akt/mTOR/p70S6K Pathway is Involved in S100A4-induced Viability and Migration in Colorectal Cancer Cells. Int. J. Med. Sci. 2014, 11, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Shubbar, E.; Vegfors, J.; Carlström, M.; Petersson, S.; Enerbäck, C. Psoriasin (S100A7) increases the expression of ROS and VEGF and acts through RAGE to promote endothelial cell proliferation. Breast Cancer Res. Treat. 2011, 134, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Hernández, J.L.; Padilla, L.; Dakhel, S.; Coll, T.; Hervas, R.; Adan, J.; Masa, M.; Mitjans, F.; Martinez, J.M.; Coma, S.; et al. Therapeutic Targeting of Tumor Growth and Angiogenesis with a Novel Anti-S100A4 Monoclonal Antibody. PLoS ONE 2013, 8, e72480. [Google Scholar] [CrossRef]
- Fuentes, M.K.; Nigavekar, S.S.; Arumugam, T.; Logsdon, C.D.; Schmidt, A.M.; Park, J.C.; Huang, E.H. RAGE Activation by S100P in Colon Cancer Stimulates Growth, Migration, and Cell Signaling Pathways. Dis. Colon Rectum 2007, 50, 1230–1240. [Google Scholar] [CrossRef]
- Hermani, A.; De Servi, B.; Medunjanin, S.; Tessier, P.A.; Mayer, D. S100A8 and S100A9 activate MAP kinase and NF-kappaB signaling pathways and trigger translocation of RAGE in human prostate cancer cells. Exp. Cell Res. 2006, 312, 184–197. [Google Scholar] [CrossRef]
- Ichikawa, M.; Williams, R.; Wang, L.; Vogl, T.; Srikrishna, G. S100A8/A9 Activate Key Genes and Pathways in Colon Tumor Progression. Mol. Cancer Res. 2011, 9, 133–148. [Google Scholar] [CrossRef]
- Li, A.; Shi, D.; Xu, B.; Wang, J.; Tang, Y.-L.; Xiao, W.; Shen, G.; Deng, W.; Zhao, C. S100A6 promotes cell proliferation in human nasopharyngeal carcinoma via the p38/MAPK signaling pathway. Mol. Carcinog. 2017, 56, 972–984. [Google Scholar] [CrossRef]
- Nasser, M.W.; Wani, N.A.; Ahirwar, D.K.; Powell, C.A.; Ravi, J.; Elbaz, M.; Zhao, H.; Padilla, L.; Zhang, X.; Shilo, K.; et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer Res. 2015, 75, 974–985. [Google Scholar] [CrossRef]
- Medapati, M.R.; Dahlmann, M.; Ghavami, S.; Pathak, K.A.; Lucman, L.; Klonisch, T.; Hoang-Vu, C.; Stein, U.; Hombach-Klonisch, S. RAGE Mediates the Pro-Migratory Response of Extracellular S100A4 in Human Thyroid Cancer Cells. Thyroid 2015, 25, 514–527. [Google Scholar] [CrossRef]
- Emberley, E.D.; Murphy, L.C.; Watson, P.H. S100A7 and the progression of breast cancer. Breast Cancer Res. 2004, 6, 153–159. [Google Scholar] [CrossRef]
- Dakhel, S.; Padilla, L.; Adan, J.; Masa, M.; Martinez, J.M.; Roque, L.; Coll, T.; Hervas, R.; Calvis, C.; Messeguer, R.; et al. S100P antibody-mediated therapy as a new promising strategy for the treatment of pancreatic cancer. Oncogenesis 2014, 3, e92. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, C.; Shan, X.; Wu, J.; Liu, H.; Liu, H.; Zhang, J.; Xu, W.; Sha, Z.; He, J.; et al. S100P is associated with proliferation and migration in nasopharyngeal carcinoma. Oncol. Lett. 2017, 14, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Fritz, G.; Weibel, M.; Heizmann, C.W.; Galichet, A. S100B and S100A6 Differentially Modulate Cell Survival by Interacting with Distinct RAGE (Receptor for Advanced Glycation End Products) Immunoglobulin Domains. J. Biol. Chem. 2007, 282, 31317–31331. [Google Scholar] [CrossRef] [PubMed]
- Wan, S.-G.; Taccioli, C.; Jiang, Y.; Chen, H.; Smalley, K.J.; Huang, K.; Liu, X.-P.; Farber, J.L.; Croce, C.M.; Fong, L.Y.Y. Zinc deficiency activates S100A8 inflammation in the absence of COX-2 and promotes murine oral-esophageal tumor progression. Int. J. Cancer 2011, 129, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Chen, H.; Luo, A.; Ding, F.; Liu, Z. S100A14 Stimulates Cell Proliferation and Induces Cell Apoptosis at Different Concentrations via Receptor for Advanced Glycation End Products (RAGE). PLoS ONE 2011, 6, e19375. [Google Scholar] [CrossRef]
- Herwig, N.; Belter, B.; Pietzsch, J. Extracellular S100A4 affects endothelial cell integrity and stimulates transmigration of A375 melanoma cells. Biochem. Biophys. Res. Commun. 2016, 477, 963–969. [Google Scholar] [CrossRef]
- Lin, J.; Blake, M.; Tang, C.; Zimmer, D.; Rustandi, R.R.; Weber, D.J.; Carrier, F. Inhibition of p53 Transcriptional Activity by the S100B Calcium-binding Protein. J. Biol. Chem. 2001, 276, 35037–35041. [Google Scholar] [CrossRef]
- Lin, J.; Yang, Q.; Yan, Z.; Markowitz, J.; Wilder, P.T.; Carrier, F.; Weber, D.J. Inhibiting S100B Restores p53 Levels in Primary Malignant Melanoma Cancer Cells. J. Biol. Chem. 2004, 279, 34071–34077. [Google Scholar] [CrossRef]
- Satyamoorthy, K.; Chehab, N.H.; Waterman, M.J.; Lien, M.C.; El-Deiry, W.S.; Herlyn, M.; Halazonetis, T.D. Aberrant regulation and function of wild-type p53 in radioresistant melanoma cells. Cell Growth Differ. 2000, 11, 467–474. [Google Scholar]
- Yang, T.; Cheng, J.; Yang, Y.; Qi, W.; Zhao, Y.; Long, H.; Xie, R.; Zhu, B. S100B Mediates Stemness of Ovarian Cancer Stem-Like Cells Through Inhibiting p53. Stem Cells 2017, 35, 325–336. [Google Scholar] [CrossRef]
- Zhu, W.; Xue, Y.; Liang, C.; Zhang, R.; Zhang, Z.; Li, H.; Su, D.; Liang, X.; Zhang, Y.; Huang, Q.; et al. S100A16 promotes cell proliferation and metastasis via AKT and ERK cell signaling pathways in human prostate cancer. Tumor Biol. 2016, 37, 12241–12250. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Costa, M. PI3K/Akt/mTOR Signaling Pathway and the Biphasic Effect of Arsenic in Carcinogenesis. Mol. Pharmacol. 2018, 94, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Siveen, K.S.; Sikka, S.; Surana, R.; Dai, X.; Zhang, J.; Kumar, A.P.; Tan, B.K.H.; Sethi, G.; Bishayee, A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim. Biophys. Acta Rev. Cancer 2014, 1845, 136–154. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Shen, L.; Yang, K.; Huang, D.; Li, X.; Li, Y.; Zhao, L.; Chen, J.; Yi, Q.; Xu, H.; et al. C6 glioma-conditioned medium induces malignant transformation of mesenchymal stem cells: Possible role of S100B/RAGE pathway. Biochem. Biophys. Res. Commun. 2018, 495, 78–85. [Google Scholar] [CrossRef]
- Zhang, F.; Banker, G.; Liu, X.; Suwanabol, P.A.; Lengfeld, J.; Yamanouchi, D.; Kent, K.C.; Liu, B.; Goldin, A.; Beckman, J.A.; et al. The novel function of advanced glycation end products in regulation of MMP-9 production. J. Surg. Res. 2011, 171, 871–876. [Google Scholar] [CrossRef]
- Furtek, S.L.; Backos, D.S.; Matheson, C.J.; Reigan, P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem. Biol. 2016, 11, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Haase-Kohn, C.; Wolf, S.; Lenk, J.; Pietzsch, J. Copper-mediated cross-linking of S100A4, but not of S100A2, results in proinflammatory effects in melanoma cells. Biochem. Biophys. Res. Commun. 2011, 413, 494–498. [Google Scholar] [CrossRef]
- Herwig, N.; Belter, B.; Wolf, S.; Haase-Kohn, C.; Pietzsch, J. Interaction of extracellular S100A4 with RAGE prompts prometastatic activation of A375 melanoma cells. J. Cell. Mol. Med. 2016, 20, 825–835. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Hua, Z.; Ma, J.; Wu, X.; Liu, Z.; Chen, H.; Cui, Z. S100A7 promotes the migration, invasion and metastasis of human cervical cancer cells through epithelial-mesenchymal transition. Oncotarget 2017, 8, 24964–24977. [Google Scholar] [CrossRef]
- Taccioli, C.; Wan, S.-G.; Liu, C.-G.; Alder, H.; Volinia, S.; Farber, J.L.; Croce, C.M.; Fong, L.Y.Y. Zinc replenishment reverses overexpression of the proinflammatory mediator S100A8 and esophageal preneoplasia in the rat. Gastroenterology 2009, 136, 953–966. [Google Scholar] [CrossRef][Green Version]
- De Ponti, A.; Wiechert, L.; Schneller, D.; Pusterla, T.; Longerich, T.; Hogg, N.; Vogel, A.; Schirmacher, P.; Hess, J.; Angel, P. A pro-tumorigenic function of S100A8/A9 in carcinogen-induced hepatocellular carcinoma. Cancer Lett. 2015, 369, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Iotzova-Weiss, G.; Dziunycz, P.J.; Freiberger, S.N.; Läuchli, S.; Hafner, J.; Vogl, T.; French, L.E.; Hofbauer, G.F.L. S100A8/A9 stimulates keratinocyte proliferation in the development of squamous cell carcinoma of the skin via the receptor for advanced glycation-end products. PLoS ONE 2015, 10, e0120971. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Duan, L.; Cui, F.; Cao, J.; Xiang, Y.; Tang, Y.; Zhou, L. S100A9 promotes human hepatocellular carcinoma cell growth and invasion through RAGE-mediated ERK1/2 and p38 MAPK pathways. Exp. Cell Res. 2015, 334, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.H.; Fuentes, M.K.; Arumugam, T.; Logsdon, C.D. The RAGE ligand, S100P, has increased expression in colon cancer. J. Am. Coll. Surg. 2004, 199, 18. [Google Scholar] [CrossRef]
- Lin, J.; Yang, Q.; Wilder, P.T.; Carrier, F.; Weber, D.J. The Calcium-binding Protein S100B Down-regulates p53 and Apoptosis in Malignant Melanoma. J. Biol. Chem. 2010, 285, 27487–27498. [Google Scholar] [CrossRef]
- Arumugam, T.; Ramachandran, V.; Logsdon, C.D. Effect of Cromolyn on S100P Interactions With RAGE and Pancreatic Cancer Growth and Invasion in Mouse Models. JNCI J. Natl. Cancer Inst. 2006, 98, 1806–1818. [Google Scholar] [CrossRef]
- Onyeagucha, B.C.; Mercado-Pimentel, M.E.; Hutchison, J.; Flemington, E.K.; Nelson, M.A. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Exp. Cell Res. 2013, 319, 2081–2090. [Google Scholar] [CrossRef]
- Wu, T.S.; Tan, C.T.; Chang, C.C.; Lin, B.R.; Lai, W.T.; Chen, S.T.; Yen-Ping Kuo, M.; Rau, C.L.; Jaw, F.S.; Chang, H.H. B-cell lymphoma/leukemia 10 promotes oral cancer progression through STAT1/ATF4/S100P signaling pathway. Oncogene 2014, 34, 1207–1219. [Google Scholar] [CrossRef]
- Mercado-Pimentel, M.E.; Onyeagucha, B.C.; Li, Q.; Pimentel, A.C.; Jandova, J.; Nelson, M.A. The S100P/RAGE signaling pathway regulates expression of microRNA-21 in colon cancer cells. FEBS Lett. 2015, 589, 2388–2393. [Google Scholar] [CrossRef]
- Hudson, B.I.; Lippman, M.E. Targeting RAGE Signaling in Inflammatory Disease. Annu. Rev. Med. 2018, 69, 349–364. [Google Scholar] [CrossRef]
- Fallon, M.T. Neuropathic pain in cancer. Br. J. Anaesth. 2013, 111, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Esumi, S.; Kitamura, Y.; Yokota-Kumasaki, H.; Ushio, S.; Yamada-Takemoto, A.; Nagai, R.; Ogawa, A.; Kawasaki, Y.; Sendo, T. Effects of magnesium oxide on the serum duloxetine concentration and antidepressant-like effects of duloxetine in rats. Biol. Pharm. Bull. 2018, 41, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, H.; Makimura, C.; Koyama, A.; Otsuka, M.; Okamoto, W.; Fujisaka, Y.; Kaneda, H.; Tsurutani, J.; Nakagawa, K. Pilot study of duloxetine for cancer patients with neuropathic pain non-responsive to pregabalin. Anticancer Res. 2012, 32, 1805–1809. [Google Scholar] [PubMed]
- Gao, H.; Zhang, I.Y.; Zhang, L.; Song, Y.; Liu, S.; Ren, H.; Liu, H.; Zhou, H.; Su, Y.; Yang, Y.; et al. S100B Suppression Alters Polarization of Infiltrating Myeloid-Derived Cells in Gliomas and Inhibits Tumor Growth. Cancer Lett. 2018, S0304-S3835, 30498–30501. [Google Scholar] [CrossRef]
- Pellegrini, L.; Xue, J.; Larson, D.; Pastorino, S.; Jube, S.; Forest, K.H.; Saad-Jube, Z.S.; Napolitano, A.; Pagano, I.; Negi, V.S.; et al. HMGB1 targeting by ethyl pyruvate suppresses malignant phenotype of human mesothelioma. Oncotarget 2017, 8, 22649–22661. [Google Scholar] [CrossRef]
- Chen, B.; Na, F.; Yang, H.; Li, R.; Li, M.; Sun, X.; Hu, B.; Huang, G.; Lan, J.; Xu, H.; et al. Ethyl pyruvate alleviates radiation-induced lung injury in mice. Biomed. Pharmacother. 2017, 92, 468–478. [Google Scholar] [CrossRef]
- Liu, Q.; Huo, Y.; Zheng, H.; Zhao, J.; Jia, L.; Wang, P. Ethyl pyruvate suppresses the growth, invasion and migration and induces the apoptosis of non-small cell lung cancer cells via the HMGB1/RAGE axis and the NF-κB/STAT3 pathway. Oncol. Rep. 2019, 42, 817–825. [Google Scholar] [CrossRef]
- Park, I.H.; Chung, S.K.; Lee, K.B.; Yoo, Y.C.; Kim, S.K.; Kim, G.S.; Song, K.S. An antioxidant hispidin from the mycelial cultures of Phellinus linteus. Arch. Pharm. Res. 2004, 27, 615–618. [Google Scholar] [CrossRef]
- Lim, J.H.; Lee, Y.M.; Park, S.R.; Kim, D.H.; Lim, B.O. Anticancer Activity of Hispidin via Reactive Oxygen Species-mediated Apoptosis in Colon Cancer Cells. Anticancer Res. 2014, 34, 4087–4094. [Google Scholar]
- Chandimali, N.; Huynh, D.L.; Jin, W.Y.; Kwon, T. Combination effects of hispidin and gemcitabine via inhibition of stemness in pancreatic cancer stem cells. Anticancer Res. 2018, 38, 3967–3975. [Google Scholar] [CrossRef]
- Voigtlaender, M.; Langer, F. Low-Molecular-Weight Heparin in Cancer Patients: Overview and Indications. Hamostaseologie 2019, 39, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Mizumoto, S.; Takahashi, J.; Sugahara, K. Receptor for advanced glycation end products (RAGE) functions as receptor for specific sulfated glycosaminoglycans, and anti-RAGE antibody or sulfated glycosaminoglycans delivered in vivo inhibit pulmonary metastasis of tumor cells. J. Biol. Chem. 2012, 287, 18985–18994. [Google Scholar] [CrossRef] [PubMed]
- Inada, M.; Shindo, M.; Kobayashi, K.; Sato, A.; Yamamoto, Y.; Akasaki, Y.; Ichimura, K.; Tanuma, S. Anticancer effects of a non-narcotic opium alkaloid medicine, papaverine, in human glioblastoma cells. PLoS ONE 2019, 14, e0216358. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Far, A.H.; Sroga, G.; Al Jaouni, S.K.; Mousa, S.A. Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 3613. https://doi.org/10.3390/ijms21103613
El-Far AH, Sroga G, Al Jaouni SK, Mousa SA. Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression. International Journal of Molecular Sciences. 2020; 21(10):3613. https://doi.org/10.3390/ijms21103613
Chicago/Turabian StyleEl-Far, Ali H., Grazyna Sroga, Soad K. Al Jaouni, and Shaker A. Mousa. 2020. "Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression" International Journal of Molecular Sciences 21, no. 10: 3613. https://doi.org/10.3390/ijms21103613
APA StyleEl-Far, A. H., Sroga, G., Al Jaouni, S. K., & Mousa, S. A. (2020). Role and Mechanisms of RAGE-Ligand Complexes and RAGE-Inhibitors in Cancer Progression. International Journal of Molecular Sciences, 21(10), 3613. https://doi.org/10.3390/ijms21103613







