Differential Secretome Profiling of Human Osteoarthritic Synoviocytes Treated with Biotechnological Unsulfated and Marine Sulfated Chondroitins
Abstract
:1. Introduction
2. Results
2.1. Secretome Profiling of the Synoviocytes Response to BC and CS Treatment
2.2. Cellular Localization of the Differentially Expressed Proteins in the OA Synoviocytes Secretome
2.3. Comparative Analysis of the OA Synoviocytes Secretome Response to BC and CS Treatments
2.4. Comparative Analysis of Differentially Regulated Proteins in OA Synoviocytes and Chondrocytes Secretomes in Response to Different Chodroitin Treatments
2.5. Targeted Cytokines Profiling by Multiplex Immunoassay
3. Discussion
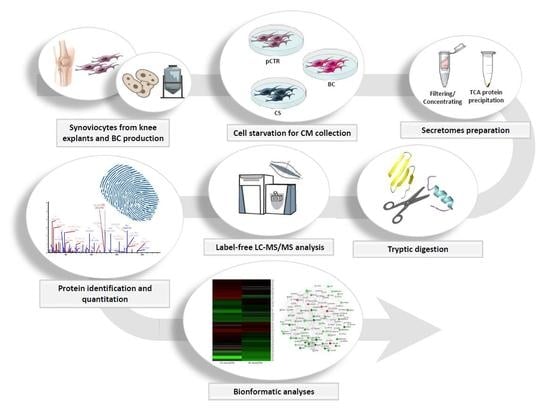
4. Materials and Methods
4.1. Preparation of CS and BC Based Gels
4.2. In Vitro Cell Cultures and Glycosaminoglycan Treatments
4.3. Targeted Quantitative Analysis of Secreted Cytokines by Bio-Plex Assay
4.4. Sample Preparation for High Resolution nanoLC-Tandem Mass Spectrometry Analyses
4.5. Database Searching and Label Free Quantitation
4.6. Bioinformatic Analyses
4.7. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Biotechnological Chondroitin |
| CS | Chondroitin Sulfate |
| ECM | Extracellular Matrix |
| FDRs | False Discovery Rates |
| GlcN | Glucosamine |
| GO | Gene Ontology |
| MS | Mass Spectrometry |
| NSAIDs | Non-Steroidal Anti-Inflammatory Drugs |
| OA | Osteoarthritis |
| SYSADOA | Symptomatic Slow-Acting Drugs |
References
- Chen, D.; Shen, J.; Zhao, W.; Wang, T.; Han, L.; Hamilton, J.L.; Im, H.J. Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 2017, 5, 16044. [Google Scholar] [CrossRef] [PubMed]
- Sellam, J.; Berenbaum, F. The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat. Rev. Rheumatol. 2010, 6, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Collin, E.C.; Carroll, O.; Kilcoyne, M.; Peroglio, M.; See, E.; Hendig, D.; Alini, M.; Grad, S.; Pandit, A. Ageing affects chondroitin sulfates and their synthetic enzymes in the intervertebral disc. Signal. Transduct Target. 2017, 2, 17049. [Google Scholar] [CrossRef] [PubMed]
- Restaino, O.F.; Finamore, R.; Stellavato, A.; Diana, P.; Bedini, E.; Trifuoggi, M.; De Rosa, M.; Schiraldi, C. European chondroitin sulfate and glucosamine food supplements: A systematic quality and quantity assessment compared to pharmaceuticals. Carbohydr. Polym. 2019, 222, 114984. [Google Scholar] [CrossRef]
- Honvo, G.; Bruyere, O.; Reginster, J.Y. Update on the role of pharmaceutical-grade chondroitin sulfate in the symptomatic management of knee osteoarthritis. Aging Clin. Exp. Res. 2019, 31, 1163–1167. [Google Scholar] [CrossRef] [Green Version]
- Cimini, D.; Rosa, M.D.; Schiraldi, C. Production of glucuronic acid-based polysaccharides by microbial fermentation for biomedical applications. Biotechnol. J. 2012, 7, 237–250. [Google Scholar] [CrossRef]
- Zhang, W.; Moskowitz, R.W.; Nuki, G.; Abramson, S.; Altman, R.D.; Arden, N.; Bierma-Zeinstra, S.; Brandt, K.D.; Croft, P.; Doherty, M.; et al. Oarsi recommendations for the management of hip and knee osteoarthritis, part ii: Oarsi evidence-based, expert consensus guidelines. Osteoarthr. Cartil. 2008, 16, 137–162. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Nuki, G.; Moskowitz, R.W.; Abramson, S.; Altman, R.D.; Arden, N.K.; Bierma-Zeinstra, S.; Brandt, K.D.; Croft, P.; Doherty, M.; et al. Oarsi recommendations for the management of hip and knee osteoarthritis: Part iii: Changes in evidence following systematic cumulative update of research published through january 2009. Osteoarthr. Cartil. 2010, 18, 476–499. [Google Scholar] [CrossRef] [Green Version]
- Hochberg, M.; Chevalier, X.; Henrotin, Y.; Hunter, D.J.; Uebelhart, D. Symptom and structure modification in osteoarthritis with pharmaceutical-grade chondroitin sulfate: What’s the evidence? Curr. Med. Res. Opin. 2013, 29, 259–267. [Google Scholar] [CrossRef]
- Henrotin, Y.; Mathy, M.; Sanchez, C.; Lambert, C. Chondroitin sulfate in the treatment of osteoarthritis: From in vitro studies to clinical recommendations. Ther. Adv. Musculoskelet. Dis. 2010, 2, 335–348. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Doherty, M.; Arden, N.; Bannwarth, B.; Bijlsma, J.; Gunther, K.P.; Hauselmann, H.J.; Herrero-Beaumont, G.; Jordan, K.; Kaklamanis, P.; et al. Eular evidence based recommendations for the management of hip osteoarthritis: Report of a task force of the eular standing committee for international clinical studies including therapeutics (escisit). Ann. Rheum. Dis. 2005, 64, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Zoppetti, G.; Oreste, P. Process for the Preparation of Chondroitin Sulfates from k4 Polysaccharide and Obtained Products. U.S. Patent No. 6,777,398, 17 August 2004. [Google Scholar]
- Rodriguez, M.L.; Jann, B.; Jann, K. Structure and serological characteristics of the capsular k4 antigen of escherichia coli o5:K4:H4, a fructose-containing polysaccharide with a chondroitin backbone. Eur. J. Biochem. 1988, 177, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Schiraldi, C.; Alfano, A.; Cimini, D.; Rosa, M.D.; Panariello, A.; Restaino, O.F.; Rosa, M.D. Application of a 22l scale membrane bioreactor and cross-flow ultrafiltration to obtain purified chondroitin. Biotechnol. Prog 2012, 28, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Volpi, N. Quality of different chondroitin sulfate preparations in relation to their therapeutic activity. J. Pharm. Pharmacol. 2009, 61, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Stellavato, A.; Tirino, V.; de Novellis, F.; Della Vecchia, A.; Cinquegrani, F.; De Rosa, M.; Papaccio, G.; Schiraldi, C. Biotechnological chondroitin a novel glycosamminoglycan with remarkable biological function on human primary chondrocytes. J. Cell. Biochem. 2016, 117, 2158–2169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratneswaran, A.; Rockel, J.S.; Kapoor, M. Understanding osteoarthritis pathogenesis: A multiomics system-based approach. Curr. Opin. Rheumatol. 2020, 32, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Severino, V.; Farina, A.; Chambery, A. Analysis of secreted proteins. Methods Mol. Biol. 2013, 1002, 37–60. [Google Scholar]
- Lourido, L.; Calamia, V.; Mateos, J.; Fernandez-Puente, P.; Fernandez-Tajes, J.; Blanco, F.J.; Ruiz-Romero, C. Quantitative proteomic profiling of human articular cartilage degradation in osteoarthritis. J. Proteome Res. 2014, 13, 6096–6106. [Google Scholar] [CrossRef]
- Polacek, M.; Bruun, J.A.; Johansen, O.; Martinez, I. Differences in the secretome of cartilage explants and cultured chondrocytes unveiled by silac technology. J. Orthop. Res. 2010, 28, 1040–1049. [Google Scholar] [CrossRef] [Green Version]
- Calamia, V.; Fernandez-Puente, P.; Mateos, J.; Lourido, L.; Rocha, B.; Montell, E.; Verges, J.; Ruiz-Romero, C.; Blanco, F.J. Pharmacoproteomic study of three different chondroitin sulfate compounds on intracellular and extracellular human chondrocyte proteomes. Mol. Cell. Proteom. 2012, 11, M111.013417. [Google Scholar] [CrossRef] [Green Version]
- Calamia, V.; Lourido, L.; Fernandez-Puente, P.; Mateos, J.; Rocha, B.; Montell, E.; Verges, J.; Ruiz-Romero, C.; Blanco, F.J. Secretome analysis of chondroitin sulfate-treated chondrocytes reveals anti-angiogenic, anti-inflammatory and anti-catabolic properties. Arthritis Res. 2012, 14, R202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calamia, V.; Ruiz-Romero, C.; Rocha, B.; Fernandez-Puente, P.; Mateos, J.; Montell, E.; Verges, J.; Blanco, F.J. Pharmacoproteomic study of the effects of chondroitin and glucosamine sulfate on human articular chondrocytes. Arthritis Res. 2010, 12, R138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calamia, V.; Mateos, J.; Fernandez-Puente, P.; Lourido, L.; Rocha, B.; Fernandez-Costa, C.; Montell, E.; Verges, J.; Ruiz-Romero, C.; Blanco, F.J. A pharmacoproteomic study confirms the synergistic effect of chondroitin sulfate and glucosamine. Sci. Rep. 2014, 4, 5069. [Google Scholar] [CrossRef] [PubMed]
- Daigo, K.; Yamaguchi, N.; Kawamura, T.; Matsubara, K.; Jiang, S.; Ohashi, R.; Sudou, Y.; Kodama, T.; Naito, M.; Inoue, K.; et al. The proteomic profile of circulating pentraxin 3 (ptx3) complex in sepsis demonstrates the interaction with azurocidin 1 and other components of neutrophil extracellular traps. Mol. Cell. Proteom. 2012, 11, M111.015073. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, C.; Bay-Jensen, A.C.; Pap, T.; Dvir-Ginzberg, M.; Quasnichka, H.; Barrett-Jolley, R.; Mobasheri, A.; Henrotin, Y. Chondrocyte secretome: A source of novel insights and exploratory biomarkers of osteoarthritis. Osteoarthr. Cartil. 2017, 25, 1199–1209. [Google Scholar] [CrossRef] [Green Version]
- Catterall, J.B.; Rowan, A.D.; Sarsfield, S.; Saklatvala, J.; Wait, R.; Cawston, T.E. Development of a novel 2d proteomics approach for the identification of proteins secreted by primary chondrocytes after stimulation by il-1 and oncostatin m. Rheumatology (Oxford) 2006, 45, 1101–1109. [Google Scholar] [CrossRef] [Green Version]
- Riffault, M.; Moulin, D.; Grossin, L.; Mainard, D.; Magdalou, J.; Vincourt, J.B. Label-free relative quantification applied to lc-maldi acquisition for rapid analysis of chondrocyte secretion modulation. J. Proteom. 2015, 114, 263–273. [Google Scholar] [CrossRef]
- Polacek, M.; Bruun, J.A.; Elvenes, J.; Figenschau, Y.; Martinez, I. The secretory profiles of cultured human articular chondrocytes and mesenchymal stem cells: Implications for autologous cell transplantation strategies. Cell Transplant. 2011, 20, 1381–1393. [Google Scholar] [CrossRef]
- Polacek, M.; Bruun, J.A.; Johansen, O.; Martinez, I. Comparative analyses of the secretome from dedifferentiated and redifferentiated adult articular chondrocytes. Cartilage 2011, 2, 186–196. [Google Scholar] [CrossRef] [Green Version]
- Haglund, L.; Bernier, S.M.; Onnerfjord, P.; Recklies, A.D. Proteomic analysis of the lps-induced stress response in rat chondrocytes reveals induction of innate immune response components in articular cartilage. Matrix Biol. 2008, 27, 107–118. [Google Scholar] [CrossRef]
- Lourido, L.; Calamia, V.; Fernandez-Puente, P.; Mateos, J.; Oreiro, N.; Blanco, F.J.; Ruiz-Romero, C. Secretome analysis of human articular chondrocytes unravels catabolic effects of nicotine on the joint. Proteom. Clin. Appl. 2016, 10, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Rocha, B.; Calamia, V.; Casas, V.; Carrascal, M.; Blanco, F.J.; Ruiz-Romero, C. Secretome analysis of human mesenchymal stem cells undergoing chondrogenic differentiation. J. Proteome Res. 2014, 13, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, J.; Ruetschi, U.; Skioldebrand, E.; Karrholm, J.; Lindahl, A. Quantitative proteomics reveals regulatory differences in the chondrocyte secretome from human medial and lateral femoral condyles in osteoarthritic patients. Proteome Sci. 2013, 11, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, D.W.; Ahmed, N.; Parreno, J.; Lunstrum, G.P.; Gross, A.E.; Diamandis, E.P.; Kandel, R.A. Collagen type xii and versican are present in the early stages of cartilage tissue formation by both redifferentating passaged and primary chondrocytes. Tissue Eng. Part. A 2015, 21, 683–693. [Google Scholar] [CrossRef]
- Calamia, V.; Rocha, B.; Mateos, J.; Fernandez-Puente, P.; Ruiz-Romero, C.; Blanco, F.J. Metabolic labeling of chondrocytes for the quantitative analysis of the interleukin-1-beta-mediated modulation of their intracellular and extracellular proteomes. J. Proteome Res. 2011, 10, 3701–3711. [Google Scholar] [CrossRef]
- Tang, S.; Deng, S.; Guo, J.; Chen, X.; Zhang, W.; Cui, Y.; Luo, Y.; Yan, Z.; He, Q.Y.; Shen, S.; et al. Deep coverage tissue and cellular proteomics revealed il-1beta can independently induce the secretion of tnf-associated proteins from human synoviocytes. J. Immunol. 2018, 200, 821–833. [Google Scholar] [CrossRef]
- Shen, S.; Guo, J.; Luo, Y.; Zhang, W.; Cui, Y.; Wang, Q.; Zhang, Z.; Wang, T. Functional proteomics revealed il-1beta amplifies tnf downstream protein signals in human synoviocytes in a tnf-independent manner. Biochem. Biophys. Res. Commun. 2014, 450, 538–544. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Bhattacharjee, M.; Ahmad, S.; Nirujogi, R.S.; Renuse, S.; Subbannayya, Y.; Marimuthu, A.; Srikanth, S.M.; Raju, R.; Dhillon, M.; et al. Differential proteomic analysis of synovial fluid from rheumatoid arthritis and osteoarthritis patients. Clin. Proteom. 2014, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Balakrishnan, L.; Nirujogi, R.S.; Ahmad, S.; Bhattacharjee, M.; Manda, S.S.; Renuse, S.; Kelkar, D.S.; Subbannayya, Y.; Raju, R.; Goel, R.; et al. Proteomic analysis of human osteoarthritis synovial fluid. Clin. Proteom. 2014, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Gobezie, R.; Kho, A.; Krastins, B.; Sarracino, D.A.; Thornhill, T.S.; Chase, M.; Millett, P.J.; Lee, D.M. High abundance synovial fluid proteome: Distinct profiles in health and osteoarthritis. Arthritis Res. 2007, 9, R36. [Google Scholar] [CrossRef] [Green Version]
- Kamphorst, J.J.; van der Heijden, R.; DeGroot, J.; Lafeber, F.P.; Reijmers, T.H.; van El, B.; Tjaden, U.R.; van der Greef, J.; Hankemeier, T. Profiling of endogenous peptides in human synovial fluid by nanolc-ms: Method validation and peptide identification. J. Proteome Res. 2007, 6, 4388–4396. [Google Scholar] [CrossRef] [PubMed]
- Ritter, S.Y.; Subbaiah, R.; Bebek, G.; Crish, J.; Scanzello, C.R.; Krastins, B.; Sarracino, D.; Lopez, M.F.; Crow, M.K.; Aigner, T.; et al. Proteomic analysis of synovial fluid from the osteoarthritic knee: Comparison with transcriptome analyses of joint tissues. Arthritis Rheumatol. 2013, 65, 981–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagiwa, H.; Sarkar, G.; Charlesworth, M.C.; McCormick, D.J.; Bolander, M.E. Two-dimensional gel electrophoresis of synovial fluid: Method for detecting candidate protein markers for osteoarthritis. J. Orthop Sci. 2003, 8, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Peffers, M.J.; Smagul, A.; Anderson, J.R. Proteomic analysis of synovial fluid: Current and potential uses to improve clinical outcomes. Expert Rev. Proteom. 2019, 16, 287–302. [Google Scholar] [CrossRef]
- Wanner, J.; Subbaiah, R.; Skomorovska-Prokvolit, Y.; Shishani, Y.; Boilard, E.; Mohan, S.; Gillespie, R.; Miyagi, M.; Gobezie, R. Proteomic profiling and functional characterization of early and late shoulder osteoarthritis. Arthritis Res. 2013, 15, R180. [Google Scholar] [CrossRef] [Green Version]
- Mathiessen, A.; Conaghan, P.G. Synovitis in osteoarthritis: Current understanding with therapeutic implications. Arthritis Res. 2017, 19, 18. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, D.; Xu, N.; Tao, W.; Zhu, R.; Sun, R.; Fan, W.; Zhang, P.; Dong, T.; Yu, L. Follistatin-like protein 1: A serum biochemical marker reflecting the severity of joint damage in patients with osteoarthritis. Arthritis Res. 2011, 13, R193. [Google Scholar] [CrossRef] [Green Version]
- Tajika, Y.; Moue, T.; Ishikawa, S.; Asano, K.; Okumo, T.; Takagi, H.; Hisamitsu, T. Influence of periostin on synoviocytes in knee osteoarthritis. Vivo 2017, 31, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Troeberg, L.; Nagase, H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 2012, 1824, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Tiaden, A.N.; Richards, P.J. The emerging roles of htra1 in musculoskeletal disease. Am. J. Pathol. 2013, 182, 1482–1488. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Liu, W.; Bemis, A.; Wang, E.; Qiu, Y.; Morris, E.A.; Flannery, C.R.; Yang, Z. Comparative proteomic characterization of articular cartilage tissue from normal donors and patients with osteoarthritis. Arthritis Rheumatol. 2007, 56, 3675–3684. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.I.; Carozza, M.; Klein, M.; Nantermet, P.; Luk, D.; Crowl, R.M. Human htra, an evolutionarily conserved serine protease identified as a differentially expressed gene product in osteoarthritic cartilage. J. Biol. Chem. 1998, 273, 34406–34412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haridas, V.; Shetty, P.; Sarathkumar, E.; Bargale, A.; Vishwanatha, J.K.; Patil, V.; Dinesh, U.S. Reciprocal regulation of pro-inflammatory annexin a2 and anti-inflammatory annexin a1 in the pathogenesis of rheumatoid arthritis. Mol. Biol. Rep. 2019, 46, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Zhu, Y.; Jia, Y.; Jiang, H.; Zheng, X.; Liu, D.; Gao, S.; Sun, M.; Hu, B.; Jiao, B.; et al. The annexin a2 promotes development in arthritis through neovascularization by amplification hedgehog pathway. PLoS ONE 2016, 11, e0150363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zreiqat, H.; Belluoccio, D.; Smith, M.M.; Wilson, R.; Rowley, L.A.; Jones, K.; Ramaswamy, Y.; Vogl, T.; Roth, J.; Bateman, J.F.; et al. S100a8 and s100a9 in experimental osteoarthritis. Arthritis Res. 2010, 12, R16. [Google Scholar] [CrossRef] [Green Version]
- Barreto, G.; Soininen, A.; Ylinen, P.; Sandelin, J.; Konttinen, Y.T.; Nordstrom, D.C.; Eklund, K.K. Soluble biglycan: A potential mediator of cartilage degradation in osteoarthritis. Arthritis Res. 2015, 17, 379. [Google Scholar] [CrossRef] [Green Version]
- Dodge, G.R.; Boesler, E.W.; Jimenez, S.A. Expression of the basement membrane heparan sulfate proteoglycan (perlecan) in human synovium and in cultured human synovial cells. Lab. Investig. 1995, 73, 649–657. [Google Scholar]
- Kaneko, H.; Ishijima, M.; Futami, I.; Tomikawa-Ichikawa, N.; Kosaki, K.; Sadatsuki, R.; Yamada, Y.; Kurosawa, H.; Kaneko, K.; Arikawa-Hirasawa, E. Synovial perlecan is required for osteophyte formation in knee osteoarthritis. Matrix Biol. 2013, 32, 178–187. [Google Scholar] [CrossRef] [Green Version]
- Papalia, R.; Vadala, G.; Torre, G.; Perna, M.; Saccone, L.; Cannata, F.; Denaro, V. The cytokinome in osteoarthritis, a new paradigm in diagnosis and prognosis of cartilage disease. J. Biol. Regul. Homeost. Agents 2016, 30, 77–83. [Google Scholar]
- Nees, T.A.; Rosshirt, N.; Zhang, J.A.; Reiner, T.; Sorbi, R.; Tripel, E.; Walker, T.; Schiltenwolf, M.; Hagmann, S.; Moradi, B. Synovial cytokines significantly correlate with osteoarthritis-related knee pain and disability: Inflammatory mediators of potential clinical relevance. J. Clin. Med. 2019, 8, 1343. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, K.; Kumar, U.; Das, S.; Chaudhuri, J.; Kumar, P.; Kanjilal, M.; Ghosh, P.; Sircar, G.; Basyal, R.K.; Kanga, U.; et al. Synovial il-9 facilitates neutrophil survival, function and differentiation of th17 cells in rheumatoid arthritis. Arthritis Res. 2018, 20, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsushima, K.; Oppenheim, J.J. Interleukin 8 and mcaf: Novel inflammatory cytokines inducible by il 1 and tnf. Cytokine 1989, 1, 2–13. [Google Scholar] [CrossRef]
- Conaghan, P.G.; Cook, A.D.; Hamilton, J.A.; Tak, P.P. Therapeutic options for targeting inflammatory osteoarthritis pain. Nat. Rev. Rheumatol. 2019, 15, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Lacey, D.; Sampey, A.; Mitchell, R.; Bucala, R.; Santos, L.; Leech, M.; Morand, E. Control of fibroblast-like synoviocyte proliferation by macrophage migration inhibitory factor. Arthritis Rheumatol. 2003, 48, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Schiraldi, C.; Cimini, D.; De Rosa, M. Production of chondroitin sulfate and chondroitin. Appl. Microbiol. Biotechnol. 2010, 87, 1209–1220. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; De Rosa, M.; Carlino, E.; Ruggiero, A.; Schiraldi, C. Homologous overexpression of rfah in e. Coli k4 improves the production of chondroitin-like capsular polysaccharide. Microb. Cell Fact. 2013, 12, 46. [Google Scholar] [CrossRef] [Green Version]
- Stellavato, A.; Vassallo, V.; La Gatta, A.; Pirozzi, A.V.A.; De Rosa, M.; Balato, G.; D’Addona, A.; Tirino, V.; Ruosi, C.; Schiraldi, C. Novel hybrid gels made of high and low molecular weight hyaluronic acid induce proliferation and reduce inflammation in an osteoarthritis in vitro model based on human synoviocytes and chondrocytes. Biomed. Res. Int. 2019, 2019, 4328219. [Google Scholar] [CrossRef] [Green Version]
- Stellavato, A.; Restaino, O.F.; Vassallo, V.; Finamore, R.; Ruosi, C.; Cassese, E.; De Rosa, M.; Schiraldi, C. Comparative analyses of pharmaceuticals or food supplements containing chondroitin sulfate: Are their bioactivities equivalent? Adv. Ther. 2019, 36, 3221–3237. [Google Scholar] [CrossRef] [Green Version]
- Miceli, M.; Dell’Aversana, C.; Russo, R.; Rega, C.; Cupelli, L.; Ruvo, M.; Altucci, L.; Chambery, A. Secretome profiling of cytokines and growth factors reveals that neuro-glial differentiation is associated with the down-regulation of chemokine ligand 2 (mcp-1/ccl2) in amniotic fluid derived-mesenchymal progenitor cells. Proteomics 2016, 16, 674–688. [Google Scholar] [CrossRef]
- Colucci-D’Amato, L.; Cicatiello, A.E.; Reccia, M.G.; Volpicelli, F.; Severino, V.; Russo, R.; Sandomenico, A.; Doti, N.; D’Esposito, V.; Formisano, P.; et al. A targeted secretome profiling by multiplexed immunoassay revealed that secreted chemokine ligand 2 (mcp-1/ccl2) affects neural differentiation in mesencephalic neural progenitor cells. Proteomics 2015, 15, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Penna, E.; Cerciello, A.; Chambery, A.; Russo, R.; Cernilogar, F.M.; Pedone, E.M.; Perrone-Capano, C.; Cappello, S.; Di Giaimo, R.; Crispino, M. Cystatin b involvement in synapse physiology of rodent brains and human cerebral organoids. Front. Mol. Neurosci. 2019, 12, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, R.; Rega, C.; Caporale, A.; Tonon, G.; Scaramuzza, S.; Selis, F.; Ruvo, M.; Chambery, A. Ultra-performance liquid chromatography/multiple reaction monitoring mass spectrometry quantification of trastuzumab in human serum by selective monitoring of a specific peptide marker from the antibody complementarity-determining regions. Rapid Commun. Mass. Spectrom. 2017, 31, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Di Giuseppe, A.M.A.; Russo, L.; Russo, R.; Ragucci, S.; Caso, J.V.; Isernia, C.; Chambery, A.; Di Maro, A. Molecular characterization of myoglobin from sciurus vulgaris meridionalis: Primary structure, kinetics and spectroscopic studies. Biochim. Biophys. Acta Proteins Proteomics 2017, 1865, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Carafa, V.; Nebbioso, A.; Cuomo, F.; Rotili, D.; Cobellis, G.; Bontempo, P.; Baldi, A.; Spugnini, E.P.; Citro, G.; Chambery, A.; et al. Rip1-hat1-sirt complex identification and targeting in treatment and prevention of cancer. Clin. Cancer Res. 2018, 24, 2886–2900. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. Panther version 14: More genomes, a new panther go-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. Upset: Visualization of intersecting sets. IEEE Trans. Vis. Comput Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef]







| Accession | Gene | Description | CS vs. pCTR | BC vs. pCTR |
|---|---|---|---|---|
| P63104 | 1433Z | 14-3-3 protein zeta/delta | 1.5 | 1.5 |
| P35555 | FBN1 | Fibrillin-1 | 1.7 | 1.5 |
| Q12841 | FSTL1 | Follistatin-related protein 1 | 4.3 | 3.8 |
| P16035 | TIMP2 | Metalloproteinase inhibitor 2 | 4.0 | 2.0 |
| Q15063 | POSTN | Periostin | 3.3 | 1.7 |
| Q92743 | HTRA1 | Serine protease HTRA1 | 2.3 | 2.0 |
| Q01995 | TAGL | transgelin | 1.7 | 2.0 |
| P06733 | ENOA | alpha-enolase | 0.2 | 0.2 |
| Q9NZT1 | CALL5 | Calmodulin-like protein 5 | 0.3 | 0.6 |
| P01040 | CYTA | Cystatin-A | 0.2 | 0.2 |
| Q08554 | DSC1 | Desmocollin-1 | 0.1 | 0.2 |
| Q02413 | DSG1 | Desmoglein-1 | 0.1 | 0.3 |
| P15924 | DESP | Desmoplakin | 0.0 | 0.2 |
| P68104 | EF1A1 | Elongation factor 1-alpha 1 | 0.3 | 0.3 |
| P04406 | G3P | glyceraldehyde-3-phosphate dehydrogenase | 0.3 | 0.3 |
| P11142 | HSP7C | Heat shock cognate 71 kDa protein | 0.4 | 0.6 |
| P04792 | HSPB1 | Heat shock protein beta-1 | 0.4 | 0.4 |
| P07355 | ANXA2 | Isoform 2 of Annexin A2 | 0.4 | 0.5 |
| P14923 | PLAK | Junction plakoglobin | 0.0 | 0.2 |
| P32119 | PRDX2 | Peroxiredoxin-2 | 0.5 | 0.5 |
| P05109 | S10A8 | Protein S100-A8 | 0.6 | 0.6 |
| P14618 | KPYM | Pyruvate kinase PKM | 0.3 | 0.3 |
| P60174 | TPIS | Triosephosphate isomerase | 0.2 | 0.5 |
| P21810 | PGS1 | biglycan | 1.9 | 0.4 |
| P12109 | CO6A1 | Collagen alpha-1(VI) chain | 1.6 | 0.6 |
| P12111 | CO6A3 | Collagen alpha-3(VI) chain | 4.4 | 0.4 |
| P07093 | GDN | Isoform 3 of Glia-derived nexin | 1.5 | 0.6 |
| P07996 | TSP1 | thrombospondin-1 | 2.9 | 0.3 |
| Q15582 | BGH3 | Transforming growth factor-beta-induced protein ig-h3 | 2.3 | 0.5 |
| P98160 | PGBM | Basement membrane-specific heparan sulfate proteoglycan core protein | 5.6 | 0.6 |
| Accession | Gene Name | Description | CS vs. pCTR | BC vs. pCTR |
|---|---|---|---|---|
| P16070 | CD44 | CD44 antigen | 2.0 | 1.0 |
| P36222 | CH3L1 | Chitinase-3-like protein 1 | 1.5 | 1.3 |
| P12110 | CO6A2 | Collagen alpha-2(VI) chain | 1.6 | 0.9 |
| P07585 | PGS2 | decorin | 2.3 | 1.3 |
| P21333 | FLNA | Filamin-A | 3.0 | N/D |
| P22692 | IBP4 | insulin-like growth factor-binding protein 4 | 2.8 | 1.3 |
| Q06033 | ITIH3 | Inter-alpha-trypsin inhibitor heavy chain H3 | 1.6 | 0.7 |
| O43852 | CALU | Isoform 3 of Calumenin | 1.5 | 1.3 |
| P12814 | ACTN1 | Isoform 4 of Alpha-actinin-1 | 1.8 | 0.7 |
| P48061 | SDF1 | Isoform Delta of Stromal cell-derived factor 1 | 1.7 | 1.0 |
| Q14767 | LTBP2 | Latent-transforming growth factor beta-binding protein 2 | 1.9 | 0.9 |
| P05121 | PAI1 | Plasminogen activator inhibitor 1 | 2.0 | 0.8 |
| Q15113 | PCOC1 | Procollagen C-endopeptidase enhancer 1 | 1.7 | 1.1 |
| P31151 | S10A7 | Protein S100-A7 | 1.5 | 1.0 |
| P50454 | SERPH | Serpin H1 | 2.8 | 0.9 |
| Q7Z7G0 | TARSH | Target of Nesh-SH3 | 1.6 | 1.1 |
| P24821 | TENA | Tenascin | 2.0 | N/D |
| P05452 | TETN | Tetranectin | 1.7 | 1.2 |
| P68363 | TBA1B | Tubulin alpha-1B chain | 2.0 | N/D |
| O76076 | WISP2 | WNT1-inducible-signaling pathway protein 2 | 3.3 | 1.0 |
| P11021 | BIP | 78 kDa glucose-regulated protein | 0.2 | N/D |
| P02461 | CO3A1 | Collagen alpha-1(III) chain | 0.4 | 0.9 |
| P09871 | C1S | Complement C1s subcomponent | 0.6 | 0.7 |
| P01034 | CYTC | Cystatin-C | 0.5 | 1.3 |
| P17936 | IBP3 | Isoform 2 of Insulin-like growth factor-binding protein 3 | 0.3 | 0.8 |
| P05155 | IC1 | Isoform 3 of Plasma protease C1 inhibitor | 0.6 | 0.8 |
| P00558 | PGK1 | phosphoglycerate kinase 1 | 0.3 | N/D |
| P02545 | LMNA | Prelamin-A/C | 0.5 | 0.8 |
| P12273 | PIP | Prolactin-inducible protein | 0.6 | 1.1 |
| P06702 | S10A9 | Protein S100-A9 | 0.5 | 1.3 |
| Q6UWP8 | SBSN | Suprabasin | 0.4 | 1.0 |
| P07226 | TPM4 | Tropomyosin alpha-4 chain | 0.4 | 1.3 |
| P25311 | ZA2G | Zinc-alpha-2-glycoprotein | 0.2 | 0.8 |
| O43707 | ACTN4 | Alpha-actinin-4 | 1.1 | 0.4 |
| P05089 | ARGI1 | Arginase-1 | N/D | 0.2 |
| P31944 | CASPE | Caspase-14 | N/D | 0.4 |
| Q99715 | COCA1 | Collagen alpha-1(XII) chain | 1.1 | 0.3 |
| Q15517 | CDSN | corneodesmosin | N/D | 0.4 |
| Q14574 | DSC3 | Desmocollin-3 | N/D | 0.2 |
| Q01469 | FABP5 | Fatty acid-binding protein, epidermal | N/D | 0.1 |
| Q08380 | LG3BP | Galectin-3-binding protein | 0.9 | 0.6 |
| P01876 | IGHA1 | Ig alpha-1 chain C region | 1.4 | 0.6 |
| P04075 | ALDOA | Isoform 2 of Fructose-bisphosphate aldolase A | 1.0 | 0.5 |
| Q96P63 | SPB12 | Isoform 2 of Serpin B12 | N/D | 0.4 |
| P00338 | LDHA | Isoform 3 of L-lactate dehydrogenase A chain | 0.7 | 0.5 |
| Q08431 | MFGM | Lactadherin | 1.3 | 0.5 |
| P30086 | PEBP1 | phosphatidylethanolamine-binding protein 1 | N/D | 0.5 |
| P29508 | SPB3 | Serpin B3 | N/D | 0.1 |
| P07858 | CATB | Cathepsin B | 0.9 | 2.1 |
| P20962 | PTMS | Parathymosin | N/D | 1.5 |
| P09486 | SPRC | Sparc | 1.5 | 2.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russo, R.; Vassallo, V.; Stellavato, A.; Valletta, M.; Cimini, D.; Pedone, P.V.; Schiraldi, C.; Chambery, A. Differential Secretome Profiling of Human Osteoarthritic Synoviocytes Treated with Biotechnological Unsulfated and Marine Sulfated Chondroitins. Int. J. Mol. Sci. 2020, 21, 3746. https://doi.org/10.3390/ijms21113746
Russo R, Vassallo V, Stellavato A, Valletta M, Cimini D, Pedone PV, Schiraldi C, Chambery A. Differential Secretome Profiling of Human Osteoarthritic Synoviocytes Treated with Biotechnological Unsulfated and Marine Sulfated Chondroitins. International Journal of Molecular Sciences. 2020; 21(11):3746. https://doi.org/10.3390/ijms21113746
Chicago/Turabian StyleRusso, Rosita, Valentina Vassallo, Antonietta Stellavato, Mariangela Valletta, Donatella Cimini, Paolo Vincenzo Pedone, Chiara Schiraldi, and Angela Chambery. 2020. "Differential Secretome Profiling of Human Osteoarthritic Synoviocytes Treated with Biotechnological Unsulfated and Marine Sulfated Chondroitins" International Journal of Molecular Sciences 21, no. 11: 3746. https://doi.org/10.3390/ijms21113746
APA StyleRusso, R., Vassallo, V., Stellavato, A., Valletta, M., Cimini, D., Pedone, P. V., Schiraldi, C., & Chambery, A. (2020). Differential Secretome Profiling of Human Osteoarthritic Synoviocytes Treated with Biotechnological Unsulfated and Marine Sulfated Chondroitins. International Journal of Molecular Sciences, 21(11), 3746. https://doi.org/10.3390/ijms21113746