Identification of Callose Synthases in Stinging Nettle and Analysis of Their Expression in Different Tissues
Abstract
:1. Introduction
2. Results and Discussion
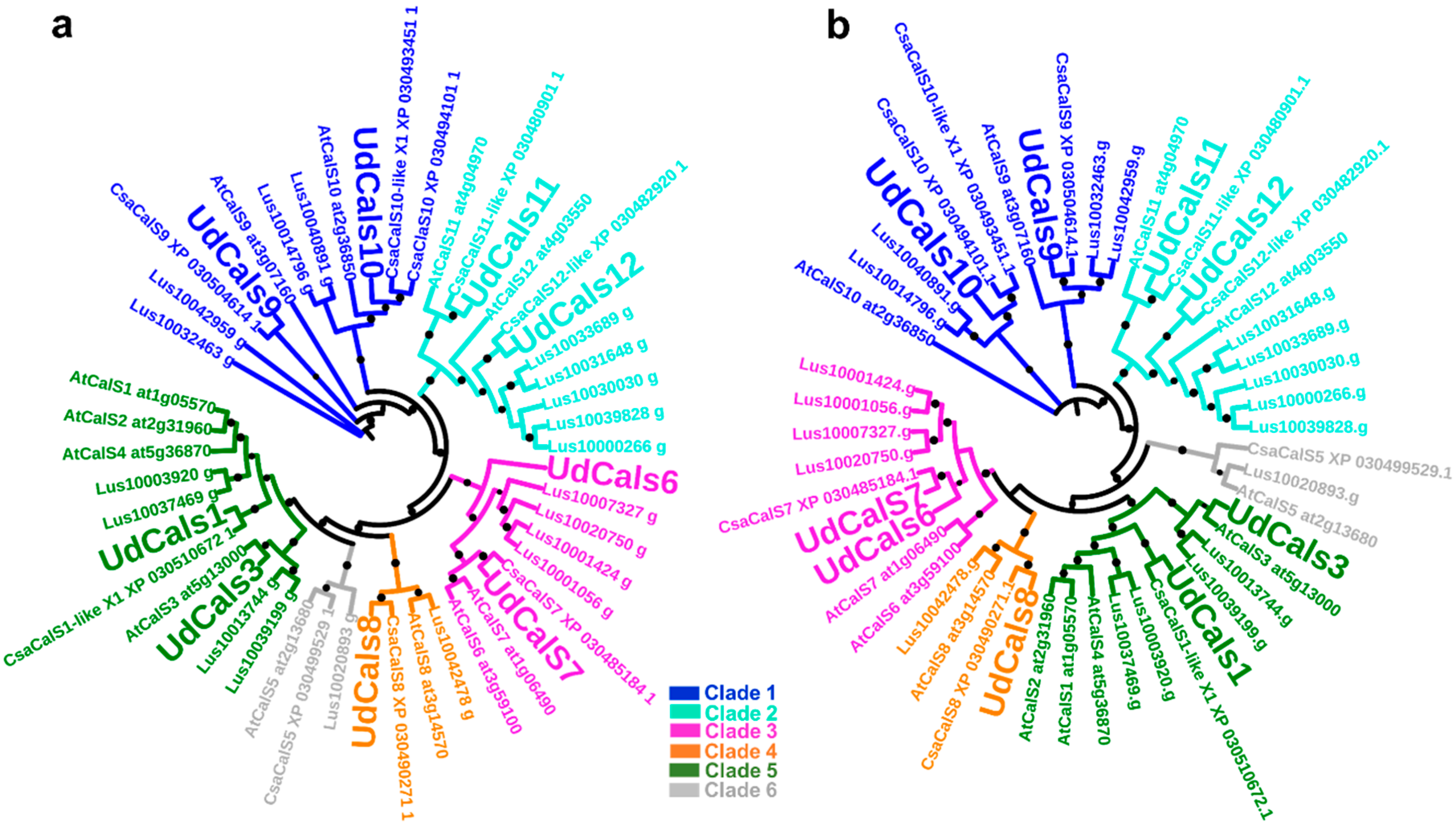
2.1. Bioinformatics and Phylogenetic Analysis of UdCalS
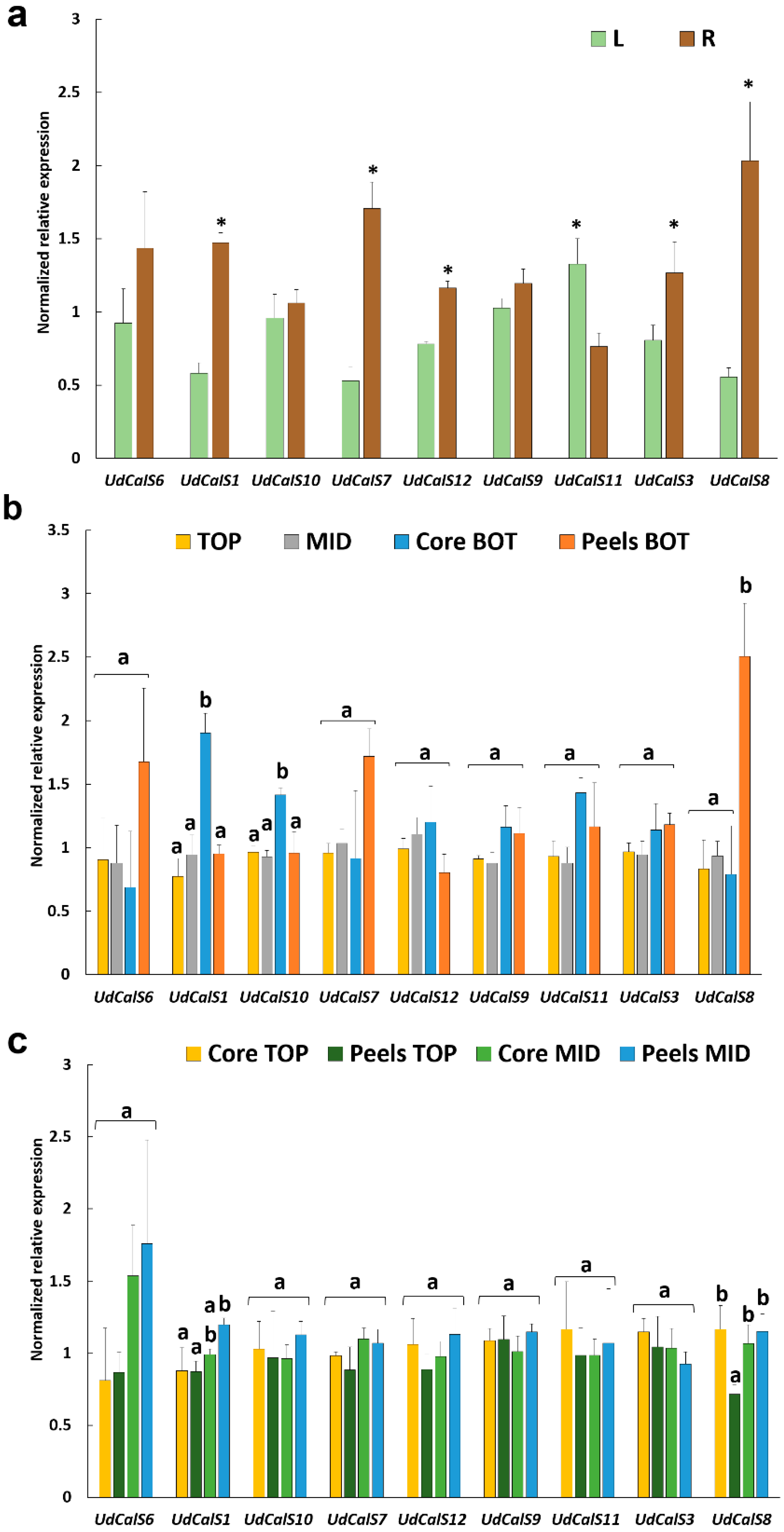
2.2. Callose Localization in Stem Tissues and Gene Expression Analysis in Different Organs
3. Materials and Methods
3.1. Plant Growth
3.2. RNA Extraction and Gene Expression Analysis
3.3. Bioinformatic Analyses
3.4. Tissue Sectioning and Confocal Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guerriero, G.; Hausman, J.-F.; Strauss, J.; Ertan, H.; Siddiqui, K.S. Lignocellulosic biomass: Biosynthesis, degradation, and industrial utilization. Eng. Life Sci. 2016, 16, 1–16. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Diffuse growth of plant cell walls. Plant Physiol. 2018, 176, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgrove, D.C.; Jarvis, M. Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant Sci. 2012, 3, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, R.; Cui, D.; Ye, Z.-H. Secondary cell wall biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerriero, G.; Sergeant, K.; Hausman, J.-F. Integrated-omics: A powerful approach to understanding the heterogeneous lignification of fibre crops. Int. J. Mol. Sci. 2013, 14, 10958–10978. [Google Scholar] [CrossRef] [Green Version]
- Guerriero, G.; Behr, M.; Legay, S.; Mangeot-Peter, L.; Zorzan, S.; Ghoniem, M.; Hausman, J.-F. Transcriptomic profiling of hemp bast fibres at different developmental stages. Sci. Rep. 2017, 7, 4961. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.-Y.; Kim, J.-Y. Callose synthesis in higher plants. Plant Signal. Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef] [Green Version]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol. Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, A.K.; Lipka, V.; Burton, R.A.; Panstruga, R.; Strizhov, N.; Schulze-Lefert, P.; Fincher, G.B. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 2003, 15, 2503–2513. [Google Scholar] [CrossRef] [Green Version]
- De Storme, N.; Geelen, D. Callose homeostasis at plasmodesmata: Molecular regulators and developmental relevance. Front. Plant Sci. 2014, 5, 138. [Google Scholar] [CrossRef] [Green Version]
- Parre, E.; Geitmann, A. More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol. 2005, 137, 274–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollet, J.-C.; Leroux, C.; Dardelle, F.; Lehner, A. Cell wall composition, biosynthesis and remodeling during pollen tube growth. Plants (Basel) 2013, 2, 107–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waterkeyn, L. Cytochemical localization and function of the 3-linked glucan callose in the developing cotton fibre cell wall. Protoplasma 1981, 106, 49–67. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Llewellyn, D.J.; Furbank, R.T. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. Plant Cell 2001, 13, 47–60. [Google Scholar] [PubMed] [Green Version]
- Rinne, P.L.; Van der Schoot, C. Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 1998, 125, 1477–1485. [Google Scholar]
- Rinne, P.L.; Kaikuranta, P.M.; Van der Schoot, C. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 2001, 26, 249–264. [Google Scholar] [CrossRef]
- Apostolakos, P.; Livanos, P.; Nikolakopoulou, T.L.; Galatis, B. The role of callose in guard-cell wall differentiation and stomatal pore formation in the fern Asplenium nidus. Ann. Bot. 2009, 104, 1373–1387. [Google Scholar] [CrossRef] [Green Version]
- Kulich, I.; Vojtíková, Z.; Glanc, M.; Ortmannová, J.; Rasmann, S.; Žárský, V. Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 2015, 168, 120–131. [Google Scholar] [CrossRef] [Green Version]
- Exley, C. A possible mechanism of biological silicification in plants. Front. Plant Sci. 2015, 6, 853. [Google Scholar] [CrossRef] [Green Version]
- Law, C.; Exley, C. New insight into silica deposition in horsetail (Equisetum arvense). BMC Plant Biol. 2011, 11, 112. [Google Scholar] [CrossRef] [Green Version]
- Guerriero, G.; Law, C.; Stokes, I.; Moore, K.L.; Exley, C. Rough and tough. How does silicic acid protect horsetail from fungal infection? J. Trace Elem. Med. Biol. 2018, 47, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Stokes, I.; Exley, C. Is callose required for silicification in plants? Biol. Lett. 2018, 14, 20180338. [Google Scholar] [CrossRef] [Green Version]
- Brugiére, T.; Exley, C. Callose-associated silica deposition in Arabidopsis. J. Trace Elem. Med. Biol. 2017, 39, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Backes, A.; Legay, S.; Berni, R.; Faleri, C.; Gatti, E.; Hausman, J.-F.; Cai, G.; Guerriero, G. Cell wall composition and transcriptomics in stem tissues of stinging nettle (Urtica dioica L.): Spotlight on a neglected fibre crop. Plant Direct 2019, 3, e00151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snegireva, A.V.; Ageeva, M.V.; Amenitskii, S.I.; Chernova, T.E.; Ebskamp, M.; Gorshkova, T.A. Intrusive growth of sclerenchyma fibers. Russ. J. Plant Physiol. 2010, 57, 342–355. [Google Scholar] [CrossRef]
- Ageeva, M.V.; Petrovská, B.; Kieft, H.; Sal’nikov, V.V.; Snegireva, A.V.; Van Dam, J.E.G.; Van Veenendaal, W.L.H.; Emons, A.M.C.; Gorshkova, T.A.; Van Lammeren, A.A.M. Intrusive growth of flax phloem fibers is of intercalary type. Planta 2005, 222, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S. Plant fibers: Initiation, growth, model plants, and open questions. Russ. J. Plant Physiol. 2010, 57, 305–315. [Google Scholar] [CrossRef]
- Gorshkova, T.; Brutch, N.; Chabbert, B.; Deyholos, M.; Hayashi, T.; Lev-Yadun, S.; Mellerowicz, E.J.; Morvan, C.; Neutelings, G.; Pilate, G. Plant fiber formation: State of the art, recent and expected progress, and open questions. Crit. Rev. Plant Sci. 2012, 31, 201–228. [Google Scholar] [CrossRef]
- Ibragimova, N.N.; Ageeva, M.V.; Gorshkova, T.A. Development of gravitropic response: Unusual behavior of flax phloem G-fibers. Protoplasma 2017, 254, 749–762. [Google Scholar] [CrossRef]
- Hong, Z.; Delauney, A.J.; Verma, D.P. A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 2001, 13, 755–768. [Google Scholar] [CrossRef] [Green Version]
- Douglas, C.M.; Foor, F.; Marrinan, J.A.; Morin, N.; Nielsen, J.B.; Dahl, A.M.; Mazur, P.; Baginsky, W.; Li, W.; El-Sherbeini, M. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc. Natl. Acad. Sci. USA 1994, 91, 12907–12911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Záveská Drábková, L.; Honys, D. Evolutionary history of callose synthases in terrestrial plants with emphasis on proteins involved in male gametophyte development. PLoS ONE 2017, 12, e0187331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Konishi, M.; Yanagisawa, S. Transcriptional repression caused by Dof5.8 is involved in proper vein network formation in Arabidopsis thaliana leaves. J. Plant Res. 2015, 128, 643–652. [Google Scholar] [CrossRef]
- Konishi, M.; Donner, T.J.; Scarpella, E.; Yanagisawa, S. MONOPTEROS directly activates the auxin-inducible promoter of the Dof5.8 transcription factor gene in Arabidopsis thaliana leaf provascular cells. J. Exp. Bot. 2015, 66, 283–291. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Hyun, T.K.; Zhang, M.; Kumar, R.; Koh, E.; Kang, B.-H.; Lucas, W.J.; Kim, J.-Y. Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev. Cell 2014, 28, 132–146. [Google Scholar] [CrossRef] [Green Version]
- Meister, R.J.; Williams, L.A.; Monfared, M.M.; Gallagher, T.L.; Kraft, E.A.; Nelson, C.G.; Gasser, C.S. Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. Plant J. 2004, 37, 426–438. [Google Scholar] [CrossRef]
- Park, D.H.; Lim, P.O.; Kim, J.S.; Cho, D.S.; Hong, S.H.; Nam, H.G. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J. 2003, 34, 161–171. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, J.; Xing, D. Phytochrome B-mediated activation of lipoxygenase modulates an excess red light-induced defence response in Arabidopsis. J. Exp. Bot. 2014, 65, 4907–4918. [Google Scholar] [CrossRef] [Green Version]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef] [Green Version]
- May, P.; Liao, W.; Wu, Y.; Shuai, B.; Richard McCombie, W.; Zhang, M.Q.; Liu, Q.A. The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 2013, 4, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Miyashima, S.; Roszak, P.; Sevilem, I.; Toyokura, K.; Blob, B.; Heo, J.-O.; Mellor, N.; Help-Rinta-Rahko, H.; Otero, S.; Smet, W.; et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skirycz, A.; Radziejwoski, A.; Busch, W.; Hannah, M.A.; Czeszejko, J.; Kwaśniewski, M.; Zanor, M.-I.; Lohmann, J.U.; De Veylder, L.; Witt, I.; et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 2008, 56, 779–792. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.-H.; Kim, S.; Lee, H.-A.; Choi, D.; Yeom, S.-I. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhao, S.; Gao, Y.; Yang, J. Characterization of Dof Transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa). PLoS ONE 2017, 12, e0170210. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Peng, H.; Lin, E.; Jin, Q.; Hua, X.; Yao, S.; Bian, H.; Han, N.; Pan, J.; Wang, J.; et al. Identification of genes related to the development of bamboo rhizome bud. J. Exp. Bot. 2010, 61, 551–561. [Google Scholar] [CrossRef] [Green Version]
- Voigt, C.A.; Schäfer, W.; Salomon, S. A comprehensive view on organ-specific callose synthesis in wheat (Triticum aestivum L.): Glucan synthase-like gene expression, callose synthase activity, callose quantification and deposition. Plant Physiol. Biochem. 2006, 44, 242–247. [Google Scholar] [CrossRef]
- Xu, X.; Guignard, C.; Renaut, J.; Hausman, J.-F.; Gatti, E.; Predieri, S.; Guerriero, G. Insights into lignan composition and biosynthesis in stinging nettle (Urtica dioica L.). Molecules 2019, 24, 3863. [Google Scholar] [CrossRef] [Green Version]
- Backes, A.; Behr, M.; Xu, X.; Gatti, E.; Legay, S.; Predieri, S.; Hausman, J.-F.; Deyholos, M.K.; Cai, G.; Guerriero, G. Sucrose synthase gene expression analysis in the fibre nettle (Urtica dioica L.) cultivar “clone 13”. Ind. Crop. Prod. 2018, 123, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; Von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Bernsel, A.; Viklund, H.; Hennerdal, A.; Elofsson, A. TOPCONS: Consensus prediction of membrane protein topology. Nucleic Acids Res. 2009, 37, W465–W468. [Google Scholar] [CrossRef] [Green Version]
- Geer, L.Y.; Domrachev, M.; Lipman, D.J.; Bryant, S.H. CDART: Protein homology by domain architecture. Genome Res. 2002, 12, 1619–1623. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; Van der Lee, R.; Bessy, A.; Chèneby, J.; Kulkarni, S.R.; Tan, G.; et al. JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–D266. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene Name with Accession Numbers | Transcript Length (nt) | Protein Length (aa) | Conserved Domains | Number of Transmembrane Domains | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| From aa | To aa | E-Value | Conserved Protein Domain Family | Short Name | TMHMM | Phobius | TOPCONS | |||
| UdCalS1 MT468368 | 5886 | 1961 | 1045 | 1805 | 0 | pfam02364 | Glucan_synthase superfamily | 16 | 17 | 19 |
| 312 | 424 | 9.97 × 10−53 | pfam14288 | FKS1_dom1 | ||||||
| 45 | 170 | 5.69 × 10−14 | pfam04652 | Vta1 superfamily | ||||||
| UdCalS3 MT468366 | 5886 | 1961 | 1059 | 1817 | 0 | pfam02364 | Glucan_synthase superfamily | 17 | 18 | 17 |
| 323 | 436 | 2.83 × 10−55 | pfam14288 | FKS1_dom1 | ||||||
| 52 | 179 | 1.78 × 10−17 | pfam04652 | Vta1 | ||||||
| UdCalS6 (partial) MT468373 | 3249 | 1082 | 216 | 961 | 0 | pfam02364 | Glucan_synthase superfamily | 10 | 10 | 11 |
| UdCalS7 MT468367 | 5778 | 1925 | 1042 | 1782 | 0 | pfam02364 | Glucan_synthase superfamily | 15 | 17 | 20 |
| 335 | 445 | 3.44 × 10−49 | pfam14288 | FKS1_dom1 | ||||||
| 56 | 191 | 4.57 × 10−11 | pfam04652 | Vta1 superfamily | ||||||
| UdCalS8 MT468374 | 5913 | 1970 | 1102 | 1834 | 0 | pfam02364 | Glucan_synthase superfamily | 13 | 17 | 17 |
| 346 | 447 | 5.48 × 10−48 | pfam14288 | FKS1_dom1 | ||||||
| 65 | 115 | 0.00046 | pfam04652 | Vta1 superfamily | ||||||
| UdCalS9 MT468369 | 5706 | 1901 | 1023 | 1766 | 0 | pfam02364 | Glucan_synthase superfamily | 16 | 17 | 17 |
| 337 | 446 | 2.45 × 10−54 | pfam14288 | FKS1_dom1 | ||||||
| UdCalS10 MT468370 | 5712 | 1903 | 1026 | 1768 | 0 | pfam02364 | Glucan_synthase superfamily | 15 | 15 | 18 |
| 347 | 457 | 4.65 × 10−52 | pfam14288 | FKS1_dom1 | ||||||
| UdCalS11 MT468372 | 5406 | 1801 | 897 | 1669 | 0 | pfam02364 | Glucan_synthase | 13 | 17 | 19 |
| 174 | 287 | 8.84 × 10−52 | pfam14288 | FKS1_dom1 | ||||||
| UdCalS12 MT468371 | 5322 | 1773 | 875 | 1639 | 0 | pfam02364 | Glucan_synthase | 12 | 18 | 19 |
| 169 | 270 | 5.78 × 10−47 | pfam14288 | FKS1_dom1 | ||||||
| CalS Clade | Conserved Motif (Significance of the Motif) | Similarity (JASPAR 2018 Plants Non-Redundant) | p-Value |
|---|---|---|---|
| 1 | TTYKTKTTTKTTYATTTTBWTTCTTWTTTTWAHKMTTWTTSTYTGHATHT (3.7 × 10−5) | MA1267.1 (AT5G66940) | 6.31 × 10−8 |
| 2 | AMACHWTCTCWHYSTCTCTCTCTCYVHC (1.6 × 10−5) | MA1402.1 (BPC6) | 3.76 × 10−11 |
| 3 | YCACAYDKCKKCHTCMTCYTSTTCTCTYTTBTVTWYCCTYT (1.5 × 10−8) TCCTCYTTMTCGCCYCGCMATGCSTTSTACTGCTWYTKCWKCYAST (9.2 × 10−8) GGCGTTAGTTTGTTCTCGTCGATCACTGCCGCTCTGGTATCAGGCGGCGG (3.6 × 10−2) | MA1279.1 (COG1) MA0553.1 (SMZ) MA0992.1 (ERF4) | 5.57 × 10−5 2.99 × 10−3 1.29 × 10−6 |
| 5 | YATBHWWBTCYCTCTCTTTYTRTTTTTNTTSWYNTNYT (6.2 × 10−10) AARDRSDTAAAGSTGAMATCTTTSBYT (7.4 × 10−4) TSVCCTTHWGKKTTTRSWTYRTTTTRAT (5.5 × 10−3) | MA1267.1 (AT5G66940) MA1404.1 (BPC1) MA1279.1 (COG1) | 3.44 × 10−7 3.72 × 10−7 1.38 × 10−3 |
| Name | Primer Sequence (5′→3′) | Tm (° C) | R2 | Efficiency (%) | Amplicon Size (bp) |
|---|---|---|---|---|---|
| UdCalS6 Fwd | TTCTCGCTCTCCGTTTCTTC | 80.21 | 0.96 | 89.32 | 82 |
| UdCalS6 Rev | ACCATCAAACCCTTGCTACG | ||||
| UdCalS1 Fwd | TCACACTCACCGAGAAAACG | 82.88 | 0.99 | 110.56 | 138 |
| UdCalS1 Rev | GCCGAAACAGAAGCTGAAAC | ||||
| UdCalS10 Fwd | TGGAAGAGGCTTTGTTGTCC | 81.81 | 0.99 | 111.60 | 146 |
| UdCalS10 Rev | AGGAAACAGGTCCGTTGTTG | ||||
| UdCalS7 Fwd | TGAGGAGGGTGAAACTTTGG | 83.01 | 0.99 | 109.73 | 149 |
| UdCalS7 Red | GGCTTGGTTGAAGAGCAAAC | ||||
| UdCalS12 Fwd | TTTGGACTCGGAGGAATCAG | 85.56 | 0.99 | 110.71 | 139 |
| UdCalS12 Rev | ATCCACGGGATGATGAAGAG | ||||
| UdCalS9 Fwd | TGTGGACAAGTTGCGAGAAG | 81.14 | 1 | 111.43 | 127 |
| UdCalS9 Rev | CCCCAAAACTTTCAGAGTCG | ||||
| UdCalS11 Fwd | TCTTGAACCAGCAGTTCGTG | 84.49 | 0.99 | 105.96 | 111 |
| UdCalS11 Rev | TCGTGAGGAAATCCCAGATG | ||||
| UdCalS3 Fwd | GCCTGATGCTCCAAAAGTTC | 79.66 | 0.99 | 109.35 | 84 |
| UdCalS3 Rev | TGCAAGGAGAAAAGGACCTC | ||||
| UdCalS8 Fwd | GTCCACGCCTTTGAAATAGC | 83.69 | 0.99 | 110.52 | 145 |
| UdCalS8 Rev | CTCGAACATCGCTCTTTTCC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerriero, G.; Piasecki, E.; Berni, R.; Xu, X.; Legay, S.; Hausman, J.-F. Identification of Callose Synthases in Stinging Nettle and Analysis of Their Expression in Different Tissues. Int. J. Mol. Sci. 2020, 21, 3853. https://doi.org/10.3390/ijms21113853
Guerriero G, Piasecki E, Berni R, Xu X, Legay S, Hausman J-F. Identification of Callose Synthases in Stinging Nettle and Analysis of Their Expression in Different Tissues. International Journal of Molecular Sciences. 2020; 21(11):3853. https://doi.org/10.3390/ijms21113853
Chicago/Turabian StyleGuerriero, Gea, Emilie Piasecki, Roberto Berni, Xuan Xu, Sylvain Legay, and Jean-Francois Hausman. 2020. "Identification of Callose Synthases in Stinging Nettle and Analysis of Their Expression in Different Tissues" International Journal of Molecular Sciences 21, no. 11: 3853. https://doi.org/10.3390/ijms21113853
APA StyleGuerriero, G., Piasecki, E., Berni, R., Xu, X., Legay, S., & Hausman, J.-F. (2020). Identification of Callose Synthases in Stinging Nettle and Analysis of Their Expression in Different Tissues. International Journal of Molecular Sciences, 21(11), 3853. https://doi.org/10.3390/ijms21113853











