Mitochondrial Inheritance in Phytopathogenic Fungi—Everything Is Known, or Is It?
Abstract
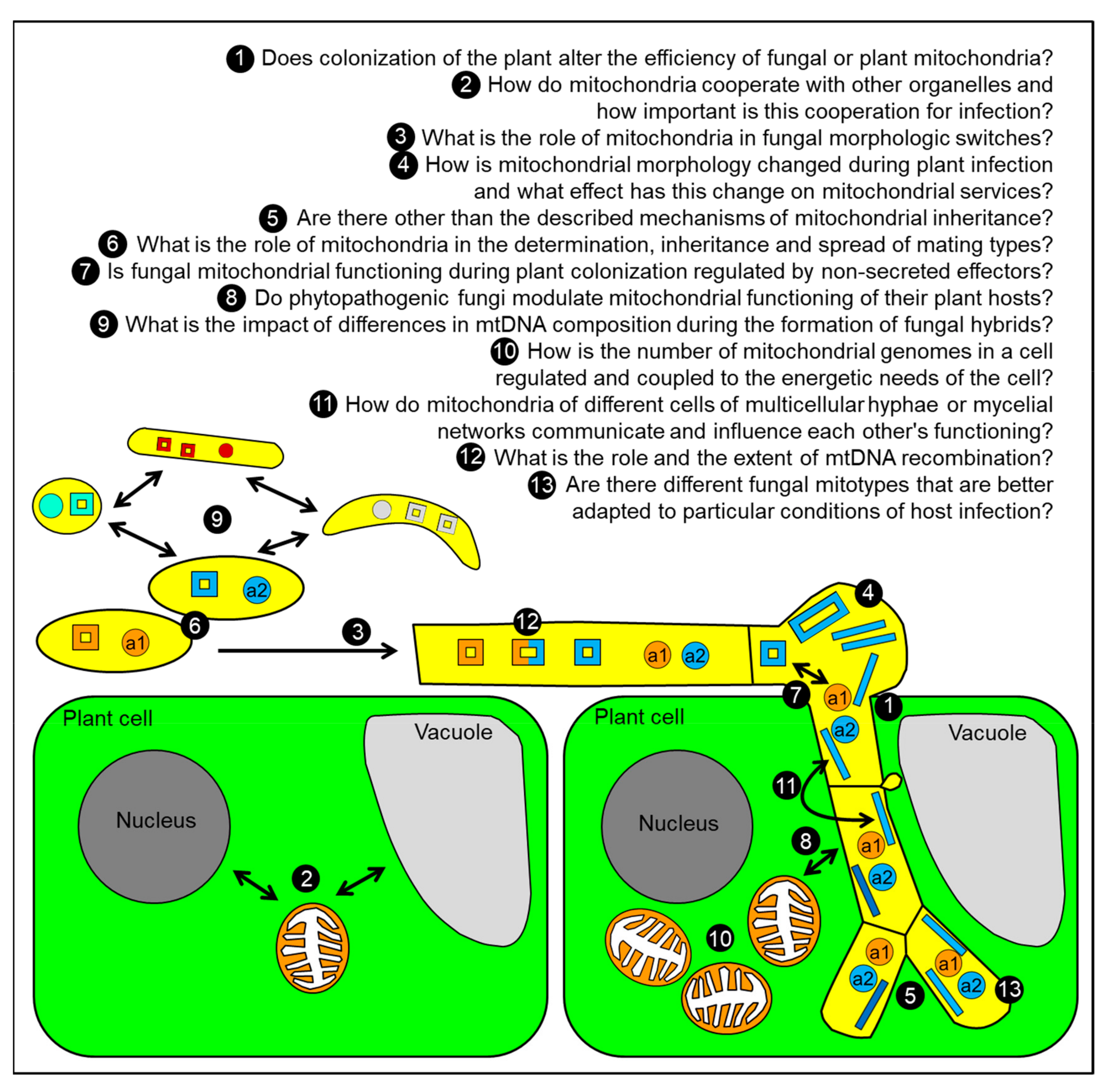
:1. Introduction
2. Mitochondria Are Critical for Cellular Energetics
3. Mitochondria Have a Long History of Cellular Co-Evolution
4. Mitochondria Vary Greatly in Number and Form
5. Mitochondrial Distribution during Cell Division Is Tightly Controlled
6. Mitochondrial Genome Stability Is Affected by Processes during and after mtDNA Replication
7. Mitochondrial Inheritance Is Differently Regulated in Different Organisms
7.1. Saccharomyces cerevisiae: Location-Dependent Mitochondrial Inheritance
7.2. Ustilago maydis: Degradation-Mediated Uniparental Mitochondrial Inheritance
7.3. Cryptococcus neoformans: Genetic and Physical Constraints during Uniparental Mitochondrial Inheritance
7.4. Microbotryum violaceum: Doubly Uniparental Inheritance of Organelles
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotiadis, V.N.; Duchen, M.R.; Osellame, L.D. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim. Biophys. Acta 2014, 1840, 1254–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christie, J.R.; Schaerf, T.M.; Beekman, M. Selection against heteroplasmy explains the evolution of uniparental inheritance of mitochondria. PLoS Genet. 2015, 11, e1005112. [Google Scholar] [CrossRef]
- Vartak, R.; Porras, C.A.; Bai, Y. Respiratory supercomplexes: Structure, function and assembly. Protein Cell 2013, 4, 582–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, M.E.; Newman, D.K. Extracellular electron transfer. Cell. Mol. Life Sci. 2001, 58, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R. Electron carriers and energy conservation in mitochondrial respiration. ChemTexts 2019, 5. [Google Scholar] [CrossRef] [Green Version]
- Bonner, W.D.; Voss, D.O. Some characteristics of mitochondria extracted from higher plants. Nature 1961, 191, 682–884. [Google Scholar]
- Ohnishi, T.; Kawaguchi, K.; Hagihara, B. Preparation and some properties of yeast mitochondria. J. Biol. Chem. 1966, 241, 1797–1806. [Google Scholar]
- Weiss, H.; von Jagow, G.; Klingenberg, M.; Bucher, T. Characterization of Neurospora crassa mitochondria prepared with a grind-mill. Eur. J. Biochem. 1970, 14, 75–82. [Google Scholar] [CrossRef]
- Buschges, R.; Bahrenberg, G.; Zimmermann, M.; Wolf, K. NADH: Ubiquinone oxidoreductase in obligate aerobic yeasts. Yeast 1994, 10, 475–479. [Google Scholar] [CrossRef]
- De Martins, V.P.; Dinamarco, T.M.; Curti, C.; Uyemura, S.A. Classical and alternative components of the mitochondrial respiratory chain in pathogenic fungi as potential therapeutic targets. J. Bioenerg. Biomembr. 2011, 43, 81–88. [Google Scholar] [CrossRef]
- Chinnery, P.F. Mitochondrial disorders overview. In GeneReviews; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Li, H.; Slone, J.; Fei, L.; Huang, T. Mitochondrial DNA variants and common diseases: A mathematical model for the diversity of age-related mtDNA mutations. Cells 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Wallace, D.C.; Chalkia, D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a021220. [Google Scholar] [CrossRef]
- Ye, K.; Lu, J.; Ma, F.; Keinan, A.; Gu, Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc. Natl. Acad. Sci. USA 2014, 111, 10654–10659. [Google Scholar] [CrossRef] [Green Version]
- Stefano, G.B.; Kream, R.M. Cancer: Mitochondrial origins. Med. Sci. Monit. 2015, 21, 3736–3739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefano, G.B.; Kream, R.M. Mitochondrial DNA heteroplasmy in human health and disease. Biomed. Rep. 2016, 4, 259–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefano, G.B.; Kream, R.M. Aging reversal and healthy longevity is in reach: Dependence on mitochondrial DNA heteroplasmy as a key molecular target. Med. Sci. Monit. 2017, 23, 2732–2735. [Google Scholar] [CrossRef] [Green Version]
- Schon, E.A.; Bonilla, E.; DiMauro, S. Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr. 1997, 29, 131–149. [Google Scholar] [CrossRef]
- Macmillan, C.; Lach, B.; Shoubridge, E.A. Variable distribution of mutant mitochondrial DNAs (tRNA(Leu[3243])) in tissues of symptomatic relatives with MELAS: The role of mitotic segregation. Neurology 1993, 43, 1586–1590. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qi, Y.; Cui, X.; Sun, Y.; Huo, Q.; Yang, Y.; Wen, X.; Tan, M.; Du, S.; Zhang, H.; et al. Heteroplasmy and copy number variations of mitochondria in 88 hepatocellular carcinoma individuals. J. Cancer 2017, 8, 4011–4017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, M.; Kumar, D. Application of mitochondrial genome information in cancer epidemiology. Clin. Chim. Acta 2007, 383, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.; Fielder, E.; Passos, J.F. Mitochondrial dysfunction and cell senescence: Deciphering a complex relationship. FEBS Lett. 2019, 593, 1566–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Victorelli, S.; Passos, J.F. Reactive oxygen species detection in senescent cells. Methods Mol. Biol. 2019, 1896, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Vasileiou, P.V.S.; Evangelou, K.; Vlasis, K.; Fildisis, G.; Panayiotidis, M.I.; Chronopoulos, E.; Passias, P.G.; Kouloukoussa, M.; Gorgoulis, V.G.; Havaki, S. Mitochondrial homeostasis and cellular senescence. Cells 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Bernhardt, D.; Hamann, A.; Osiewacz, H.D. The role of mitochondria in fungal aging. Curr. Opin. Microbiol. 2014, 22, 1–7. [Google Scholar] [CrossRef]
- Chrobok, D.; Law, S.R.; Brouwer, B.; Linden, P.; Ziolkowska, A.; Liebsch, D.; Narsai, R.; Szal, B.; Moritz, T.; Rouhier, N.; et al. Dissecting the metabolic role of mitochondria during developmental leaf senescence. Plant. Physiol. 2016, 172, 2132–2153. [Google Scholar] [CrossRef] [Green Version]
- Goldring, E.S.; Grossman, L.I.; Marmur, J. Petite mutation in yeast. II. Isolation of mutants containing mitochondrial deoxyribonucleic acid of reduced size. J. Bacteriol. 1971, 107, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 1967, 14, 255–274. [Google Scholar] [CrossRef]
- Thrash, J.C.; Boyd, A.; Huggett, M.J.; Grote, J.; Carini, P.; Yoder, R.J.; Robbertse, B.; Spatafora, J.W.; Rappe, M.S.; Giovannoni, S.J. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 2011, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wu, M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci. Rep. 2015, 5, 7949. [Google Scholar] [CrossRef] [Green Version]
- The Genetic Codes. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi (accessed on 19 July 2019).
- Gabaldon, T.; Huynen, M.A. From endosymbiont to host-controlled organelle: The hijacking of mitochondrial protein synthesis and metabolism. PLoS Comput. Biol. 2007, 3, e219. [Google Scholar] [CrossRef] [PubMed]
- Sassera, D.; Lo, N.; Epis, S.; D’Auria, G.; Montagna, M.; Comandatore, F.; Horner, D.; Pereto, J.; Luciano, A.M.; Franciosi, F.; et al. Phylogenomic evidence for the presence of a flagellum and cbb(3) oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 2011, 28, 3285–3296. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, M. Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS ONE 2014, 9, e110685. [Google Scholar] [CrossRef]
- Tyagi, S.; Pande, V.; Das, A. Whole mitochondrial genome sequence of an Indian Plasmodium falciparum field isolate. Korean J. Parasitol. 2014, 52, 99–103. [Google Scholar] [CrossRef]
- Wu, Z.; Cuthbert, J.M.; Taylor, D.R.; Sloan, D.B. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc. Natl. Acad. Sci. USA 2015, 112, 10185–10191. [Google Scholar] [CrossRef] [Green Version]
- Karnkowska, A.; Vacek, V.; Zubacova, Z.; Treitli, S.C.; Petrzelkova, R.; Eme, L.; Novak, L.; Zarsky, V.; Barlow, L.D.; Herman, E.K.; et al. A eukaryote without a mitochondrial organelle. Curr. Biol. 2016, 26, 1274–1284. [Google Scholar] [CrossRef]
- Haig, D. Intracellular evolution of mitochondrial DNA (mtDNA) and the tragedy of the cytoplasmic commons. Bioessays 2016, 38, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Hirose, M.; Schilf, P.; Gupta, Y.; Zarse, K.; Kunstner, A.; Fahnrich, A.; Busch, H.; Yin, J.; Wright, M.N.; Ziegler, A.; et al. Low-level mitochondrial heteroplasmy modulates DNA replication, glucose metabolism and lifespan in mice. Sci. Rep. 2018, 8, 5872. [Google Scholar] [CrossRef] [Green Version]
- James, A.C.; Ballard, J.W. Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 2003, 164, 187–194. [Google Scholar]
- Smigrodzki, R.M.; Khan, S.M. Mitochondrial microheteroplasmy and a theory of aging and age-related disease. Rejuvenation Res. 2005, 8, 172–198. [Google Scholar] [CrossRef]
- Yahalomi, D.; Atkinson, S.D.; Neuhof, M.; Chang, E.S.; Philippe, H.; Cartwright, P.; Bartholomew, J.L.; Huchon, D. A cnidarian parasite of salmon (Myxozoa: Henneguya) lacks a mitochondrial genome. Proc. Natl. Acad. Sci. USA 2020, 117, 5358–5363. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.W. The evolution of per-cell organelle number. Front. Cell. Dev. Biol. 2016, 4, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, L.; Bousquet, J.; Levesque, R.C.; Lalonde, M. Origin and diversification of endomycorrhizal fungi and coincidence with vascular land plants. Nature 1993, 363, 67–69. [Google Scholar] [CrossRef]
- Remy, W.; Taylor, T.N.; Hass, H.; Kerp, H. Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. USA 1994, 91, 11841–11843. [Google Scholar] [CrossRef] [Green Version]
- Nagahashi, G.; Douds, D.D., Jr. Rapid and sensitive bioassay to study signals between root exudates and arbuscular mycorrhizal fungi. Biotechnol. Techn. 1999, 13, 893–897. [Google Scholar] [CrossRef]
- Buee, M.; Rossignol, M.; Jauneau, A.; Ranjeva, R.; Becard, G. The pre-symbiotic growth of arbuscular mycorrhizal fungi is induced by a branching factor partially purified from plant root exudates. Mol. Plant Microbe Interact. 2000, 13, 693–698. [Google Scholar] [CrossRef] [Green Version]
- Tamasloukht, M.; Sejalon-Delmas, N.; Kluever, A.; Jauneau, A.; Roux, C.; Becard, G.; Franken, P. Root factors induce mitochondrial-related gene expression and fungal respiration during the developmental switch from asymbiosis to presymbiosis in the arbuscular mycorrhizal fungus Gigaspora rosea. Plant Physiol. 2003, 131, 1468–1478. [Google Scholar] [CrossRef] [Green Version]
- Bouwmeester, H.J.; Matusova, R.; Zhongkui, S.; Beale, M.H. Secondary metabolite signalling in host-parasitic plant interactions. Curr. Opin. Plant Biol. 2003, 6, 358–364. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Besserer, A.; Puech-Pages, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.C.; Roux, C.; Becard, G.; Sejalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, e226. [Google Scholar] [CrossRef]
- Besserer, A.; Becard, G.; Jauneau, A.; Roux, C.; Sejalon-Delmas, N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 2008, 148, 402–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dor, E.; Joel, D.M.; Kapulnik, Y.; Koltai, H.; Hershenhorn, J. The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta 2011, 234, 419–427. [Google Scholar] [CrossRef]
- Belmondo, S.; Marschall, R.; Tudzynski, P.; Lopez Raez, J.A.; Artuso, E.; Prandi, C.; Lanfranco, L. Identification of genes involved in fungal responses to strigolactones using mutants from fungal pathogens. Curr. Genet. 2017, 63, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Lackner, L.L. Shaping the dynamic mitochondrial network. BMC Biol. 2014, 12, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bui, H.T.; Karren, M.A.; Bhar, D.; Shaw, J.M. A novel motif in the yeast mitochondrial dynamin Dnm1 is essential for adaptor binding and membrane recruitment. J. Cell. Biol. 2012, 199, 613–622. [Google Scholar] [CrossRef] [Green Version]
- Mears, J.A.; Lackner, L.L.; Fang, S.; Ingerman, E.; Nunnari, J.; Hinshaw, J.E. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011, 18, 20–26. [Google Scholar] [CrossRef]
- Lackner, L.L.; Horner, J.S.; Nunnari, J. Mechanistic analysis of a dynamin effector. Science 2009, 325, 874–877. [Google Scholar] [CrossRef]
- Tieu, Q.; Okreglak, V.; Naylor, K.; Nunnari, J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell. Biol. 2002, 158, 445–452. [Google Scholar] [CrossRef] [Green Version]
- Koirala, S.; Bui, H.T.; Schubert, H.L.; Eckert, D.M.; Hill, C.P.; Kay, M.S.; Shaw, J.M. Molecular architecture of a dynamin adaptor: Implications for assembly of mitochondrial fission complexes. J. Cell. Biol. 2010, 191, 1127–1139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerveny, K.L.; Studer, S.L.; Jensen, R.E.; Sesaki, H. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell. 2007, 12, 363–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammermeister, M.; Schodel, K.; Westermann, B. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell. 2010, 21, 2443–2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otera, H.; Wang, C.; Cleland, M.M.; Setoguchi, K.; Yokota, S.; Youle, R.J.; Mihara, K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell. Biol. 2010, 191, 1141–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, R.; Jin, S.B.; Lendahl, U.; Nister, M.; Zhao, J. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Dorn, G.W., II. Mitofusins as mitochondrial anchors and tethers. [published online ahead of print, 2020 Apr 15]. J. Mol. Cell. Cardiol. 2020, S0022-2828(29)30099-7. [Google Scholar] [CrossRef]
- Del Dotto, V.; Fogazza, M.; Carelli, V.; Rugolo, M.; Zanna, C. Eight human OPA1 isoforms, long and short: What are they for? Biochim. Biophys. Acta Bioenerg. 2018, 1859, 263–269. [Google Scholar] [CrossRef]
- Mitra, S.; Elliott, S.J. Oxidative disassembly of the [2Fe-2S] cluster of human Grx2 and redox regulation in the mitochondria. Biochemistry 2009, 48, 3813–3815. [Google Scholar] [CrossRef]
- Gerstenberger, J.P.; Occhipinti, P.; Gladfelter, A.S. Heterogeneity in mitochondrial morphology and membrane potential is independent of the nuclear division cycle in multinucleate fungal cells. Eukaryot. Cell. 2012, 11, 353–367. [Google Scholar] [CrossRef] [Green Version]
- Boldogh, I.R.; Pon, L.A. Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta 2006, 1763, 450–462. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.C.; Palazzo, A.; Swayne, T.C.; Pon, L.A. A retention mechanism for distribution of mitochondria during cell division in budding yeast. Curr. Biol. 1999, 9, 1111–1114. [Google Scholar] [CrossRef] [Green Version]
- Rafelski, S.M.; Viana, M.P.; Zhang, Y.; Chan, Y.H.; Thorn, K.S.; Yam, P.; Fung, J.C.; Li, H.; Costa Lda, F.; Marshall, W.F. Mitochondrial network size scaling in budding yeast. Science 2012, 338, 822–824. [Google Scholar] [CrossRef] [Green Version]
- Longo, V.D.; Shadel, G.S.; Kaeberlein, M.; Kennedy, B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012, 16, 18–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFaline-Figueroa, J.R.; Vevea, J.; Swayne, T.C.; Zhou, C.; Liu, C.; Leung, G.; Boldogh, I.R.; Pon, L.A. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell 2011, 10, 885–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, E.J.; Boucher, E.; Mandato, C.A. Mitochondria-cytoskeleton associations in mammalian cytokinesis. Cell. Div. 2016, 11, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackner, L.L. The expanding and unexpected functions of mitochondria contact sites. Trends Cell. Biol. 2019, 29, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Chan, D.C. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell. Biol. 2014, 15, 634–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanki, T.; Klionsky, D.J. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008, 283, 32386–32393. [Google Scholar] [CrossRef] [Green Version]
- Koehler, C.M.; Lindberg, G.L.; Brown, D.R.; Beitz, D.C.; Freeman, A.E.; Mayfield, J.E.; Myers, A.M. Replacement of bovine mitochondrial DNA by a sequence variant within one generation. Genetics 1991, 129, 247–255. [Google Scholar]
- Krasich, R.; Copeland, W.C. DNA polymerases in the mitochondria: A critical review of the evidence. Front. Biosci. (Landmark Ed.) 2017, 22, 692–709. [Google Scholar] [CrossRef] [Green Version]
- Falkenberg, M. Mitochondrial DNA replication in mammalian cells: Overview of the pathway. Essays Biochem. 2018, 62, 287–296. [Google Scholar] [CrossRef]
- Macao, B.; Uhler, J.P.; Siibak, T.; Zhu, X.; Shi, Y.; Sheng, W.; Olsson, M.; Stewart, J.B.; Gustafsson, C.M.; Falkenberg, M. The exonuclease activity of DNA polymerase gamma is required for ligation during mitochondrial DNA replication. Nat. Commun. 2015, 6, 7303. [Google Scholar] [CrossRef] [Green Version]
- Nicholls, T.J.; Nadalutti, C.A.; Motori, E.; Sommerville, E.W.; Gorman, G.S.; Basu, S.; Hoberg, E.; Turnbull, D.M.; Chinnery, P.F.; Larsson, N.G.; et al. Topoisomerase 3alpha is required for decatenation and segregation of human mtDNA. Mol. Cell. 2018, 69, 9–23.e26. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.J.; Clark-Walker, G.D. Unveiling the mystery of mitochondrial DNA replication in yeasts. Mitochondrion 2018, 38, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Kukat, C.; Wurm, C.A.; Spahr, H.; Falkenberg, M.; Larsson, N.G.; Jakobs, S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. USA 2011, 108, 13534–13539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kukat, C.; Davies, K.M.; Wurm, C.A.; Spahr, H.; Bonekamp, N.A.; Kuhl, I.; Joos, F.; Polosa, P.L.; Park, C.B.; Posse, V.; et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. USA 2015, 112, 11288–11293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, S.C.; Uchiyama, L.F.; Nunnari, J. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 2016, 353, aaf5549. [Google Scholar] [CrossRef] [Green Version]
- Longley, M.J.; Nguyen, D.; Kunkel, T.A.; Copeland, W.C. The fidelity of human DNA polymerase gamma with and without exonucleolytic proofreading and the p55 accessory subunit. J. Biol. Chem. 2001, 276, 38555–38562. [Google Scholar] [CrossRef] [Green Version]
- Torriani, S.F.; Penselin, D.; Knogge, W.; Felder, M.; Taudien, S.; Platzer, M.; McDonald, B.A.; Brunner, P.C. Comparative analysis of mitochondrial genomes from closely related Rhynchosporium species reveals extensive intron invasion. Fungal Genet. Biol. 2014, 62, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Nissanka, N.; Moraes, C.T. Mitochondrial DNA damage and reactive oxygen species in neurodegenerative disease. FEBS Lett. 2018, 592, 728–742. [Google Scholar] [CrossRef]
- Losada, L.; Pakala, S.B.; Fedorova, N.D.; Joardar, V.; Shabalina, S.A.; Hostetler, J.; Pakala, S.M.; Zafar, N.; Thomas, E.; Rodriguez-Carres, M.; et al. Mobile elements and mitochondrial genome expansion in the soil fungus and potato pathogen Rhizoctonia solani AG-3. FEMS Microbiol. Lett. 2014, 352, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Waymire, K.G.; Narula, N.; Li, P.; Rocher, C.; Coskun, P.E.; Vannan, M.A.; Narula, J.; Macgregor, G.R.; Wallace, D.C. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science 2008, 319, 958–962. [Google Scholar] [CrossRef] [Green Version]
- Rossignol, R.; Faustin, B.; Rocher, C.; Malgat, M.; Mazat, J.P.; Letellier, T. Mitochondrial threshold effects. Biochem. J. 2003, 370, 751–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radzvilavicius, A.L.; Lane, N.; Pomiankowski, A. Sexual conflict explains the extraordinary diversity of mechanisms regulating mitochondrial inheritance. BMC Biol. 2017, 15, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burr, S.P.; Pezet, M.; Chinnery, P.F. Mitochondrial DNA heteroplasmy and purifying selection in the mammalian female germ line. Dev. Growth Differ. 2018, 60, 21–32. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Biparental inheritance of mitochondrial DNA in humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039–13044. [Google Scholar] [CrossRef] [Green Version]
- Crow, J.F. An advantage of sexual reproduction in a rapidly changing environment. J. Hered. 1992, 83, 169–173. [Google Scholar] [CrossRef]
- Hagemann, R. The foundation of extranuclear inheritance: Plastid and mitochondrial genetics. Mol. Genet. Genom. 2010, 283, 199–209. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.; Sodmergen. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant. Cell. Physiol. 2003, 44, 941–951. [Google Scholar] [CrossRef] [Green Version]
- Neale, D.B.; Marshall, K.A.; Sederoff, R.R. Chloroplast and mitochondrial DNA are paternally inherited in Sequoia sempervirens D. Don Endl. Proc. Natl. Acad. Sci. USA 1989, 86, 9347–9349. [Google Scholar] [CrossRef] [Green Version]
- Bell, G. The evolution of anisogamy. J. Theor. Biol. 1978, 73, 247–270. [Google Scholar] [CrossRef]
- DeLuca, S.Z.; O’Farrell, P.H. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell. 2012, 22, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, Y.; Yoshinari, T.; Naruse, K.; Yamada, T.; Sumi, K.; Mitani, H.; Higashiyama, T.; Kuroiwa, T. Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc. Natl. Acad. Sci. USA 2006, 103, 1382–1387. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.M.; Ge, Z.J.; Wang, Z.W.; Jiang, Z.Z.; Wang, Z.B.; Ouyang, Y.C.; Hou, Y.; Schatten, H.; Sun, Q.Y. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 13038–13043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderegg, R.J.; Betz, R.; Carr, S.A.; Crabb, J.W.; Duntze, W. Structure of Saccharomyces cerevisiae mating hormone a-factor. Identification of S-farnesyl cysteine as a structural component. J. Biol. Chem. 1988, 263, 18236–18240. [Google Scholar] [PubMed]
- Hagen, D.C.; Bruhn, L.; Westby, C.A.; Sprague, G.F., Jr. Transcription of alpha-specific genes in Saccharomyces cerevisiae: DNA sequence requirements for activity of the coregulator alpha 1. Mol. Cell. Biol. 1993, 13, 6866–6875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, S.J.; Wolfe, K.H. An evolutionary perspective on yeast mating-type switching. Genetics 2017, 206, 9–32. [Google Scholar] [CrossRef] [Green Version]
- Maddox, P.; Chin, E.; Mallavarapu, A.; Yeh, E.; Salmon, E.D.; Bloom, K. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J. Cell. Biol. 1999, 144, 977–987. [Google Scholar] [CrossRef] [Green Version]
- Rine, J.; Sprague, G.F., Jr.; Herskowitz, I. rme1 Mutation of Saccharomyces cerevisiae: Map position and bypass of mating type locus control of sporulation. Mol. Cell. Biol. 1981, 1, 958–960. [Google Scholar] [CrossRef] [Green Version]
- Strausberg, R.L.; Perlman, P.S. The effect of zygotic bud position on the transmission of mitochondrial genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 1978, 163, 131–144. [Google Scholar] [CrossRef]
- Wilson, A.J.; Xu, J. Mitochondrial inheritance: Diverse patterns and mechanisms with an emphasis on fungi. Mycology 2012, 3, 158–166. [Google Scholar] [CrossRef]
- Hewitt, S.K.; Duangrattanalert, K.; Burgis, T.; Zeef, L.A.H.; Delneri, D. Plasticity of mitochondrial DNA inheritance and its impact on nuclear gene transcription in yeast hybrids. bioRxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Van Dyck, E.; Clayton, D.A. Transcription-dependent DNA transactions in the mitochondrial genome of a yeast hypersuppressive petite mutant. Mol. Cell. Biol. 1998, 18, 2976–2985. [Google Scholar] [CrossRef] [Green Version]
- Gillissen, B.; Bergemann, J.; Sandmann, C.; Schroeer, B.; Bölker, M.; Kahmann, R. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 1992, 68, 647–657. [Google Scholar] [CrossRef] [Green Version]
- Kronstad, J.W.; Leong, S.A. The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 1990, 4, 1384–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barr, C.M.; Neiman, M.; Taylor, D.R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005, 168, 39–50. [Google Scholar] [CrossRef]
- Fedler, M.; Luh, K.S.; Stelter, K.; Nieto-Jacobo, F.; Basse, C.W. The a2 mating-type locus genes lga2 and rga2 direct uniparental mitochondrial DNA (mtDNA) inheritance and constrain mtDNA recombination during sexual development of Ustilago maydis. Genetics 2009, 181, 847–860. [Google Scholar] [CrossRef] [Green Version]
- Urban, M.; Kahmann, R.; Bölker, M. The biallelic a mating type locus of Ustilago maydis: Remnants of an additional pheromone gene indicate evolution from a multiallelic ancestor. Mol. Gen. Genet. 1996, 250, 414–420. [Google Scholar] [CrossRef]
- Mahlert, M.; Vogler, C.; Stelter, K.; Hause, G.; Basse, C.W. The a2 mating-type-locus gene lga2 of Ustilago maydis interferes with mitochondrial dynamics and fusion, partially in dependence on a Dnm1-like fission component. J. Cell. Sci. 2009, 122, 2402–2412. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Sathananthan, A.H. Early penetration of human sperm through the vestments of human eggs in vitro. Arch. Androl. 1986, 16, 183–197. [Google Scholar] [CrossRef]
- Shalgi, R.; Magnus, A.; Jones, R.; Phillips, D.M. Fate of sperm organelles during early embryogenesis in the rat. Mol. Reprod. Dev. 1994, 37, 264–271. [Google Scholar] [CrossRef]
- Kaneda, H.; Hayashi, J.; Takahama, S.; Taya, C.; Lindahl, K.F.; Yonekawa, H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 4542–4546. [Google Scholar] [CrossRef] [Green Version]
- Al Rawi, S.; Louvet-Vallee, S.; Djeddi, A.; Sachse, M.; Culetto, E.; Hajjar, C.; Boyd, L.; Legouis, R.; Galy, V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011, 334, 1144–1147. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia 1976, 68, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Lengeler, K.B.; Fox, D.S.; Fraser, J.A.; Allen, A.; Forrester, K.; Dietrich, F.S.; Heitman, J. Mating-type locus of Cryptococcus neoformans: A step in the evolution of sex chromosomes. Eukaryot. Cell. 2002, 1, 704–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hull, C.M.; Boily, M.J.; Heitman, J. Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell. 2005, 4, 526–535. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Xu, J. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics 2003, 163, 1315–1325. [Google Scholar]
- Yan, Z.; Hull, C.M.; Sun, S.; Heitman, J.; Xu, J. The mating type-specific homeodomain genes SXI1alpha and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Genet. 2007, 51, 187–195. [Google Scholar] [CrossRef]
- Hsueh, Y.P.; Fraser, J.A.; Heitman, J. Transitions in sexuality: Recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot. Cell. 2008, 7, 1847–1855. [Google Scholar] [CrossRef] [Green Version]
- Gyawali, R.; Lin, X. Prezygotic and postzygotic control of uniparental mitochondrial DNA inheritance in Cryptococcus neoformans. mBio 2013, 4, e00112–e00113. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhai, B.; Lin, X. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 2012, 8, e1002765. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Fu, C.; Ianiri, G.; Heitman, J. The pheromone and pheromone receptor mating-type locus is involved in controlling uniparental mitochondrial inheritance in Cryptococcus. Genetics 2020, 214, 703–717. [Google Scholar] [CrossRef]
- Hintz, W.; Anderson, J.B.; Horgen, P.A. Nuclear migration and mitochondrial inheritance in the mushroom Agaricus bitorquis. Genetics 1988, 119, 35–41. [Google Scholar]
- May, G.; Taylor, J.W. Patterns of mating and mitochondrial DNA inheritance in the agaric Basidiomycete Coprinus cinereus. Genetics 1988, 118, 213–220. [Google Scholar] [PubMed]
- Yan, Z.; Li, Z.; Yan, L.; Yu, Y.; Cheng, Y.; Chen, J.; Liu, Y.; Gao, C.; Zeng, L.; Sun, X.; et al. Deletion of the sex-determining gene SXI1alpha enhances the spread of mitochondrial introns in Cryptococcus neoformans. Mob. DNA 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Kemler, M.; Goker, M.; Oberwinkler, F.; Begerow, D. Implications of molecular characters for the phylogeny of the Microbotryaceae (Basidiomycota: Urediniomycetes). BMC Evol. Biol. 2006, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Yockteng, R.; Marthey, S.; Chiapello, H.; Gendrault, A.; Hood, M.E.; Rodolphe, F.; Devier, B.; Wincker, P.; Dossat, C.; Giraud, T. Expressed sequences tags of the anther smut fungus, Microbotryum violaceum, identify mating and pathogenicity genes. BMC Genomics 2007, 8, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Petit, E.; Hood, M.E. Variation in mate-recognition pheromones of the fungal genus Microbotryum. Heredity (Edinburgh) 2016, 116, 44–51. [Google Scholar] [CrossRef] [Green Version]
- Wilch, G.; Ward, S.; Castle, A. Transmission of mitochondrial DNA in Ustilago violacea. Curr. Genet. 1992, 22, 135–140. [Google Scholar] [CrossRef]
- Soroka, M. Doubly uniparental inheritance of mitochondrial DNA in the freshwater bivalve Anodonta woodiana (Bivalvia: Unionidae). Folia Biol. (Krakow) 2008, 56, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Milani, L.; Ghiselli, F.; Maurizii, M.G.; Passamonti, M. Doubly uniparental inheritance of mitochondria as a model system for studying germ line formation. PLoS ONE 2011, 6, e28194. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendoza, H.; Perlin, M.H.; Schirawski, J. Mitochondrial Inheritance in Phytopathogenic Fungi—Everything Is Known, or Is It? Int. J. Mol. Sci. 2020, 21, 3883. https://doi.org/10.3390/ijms21113883
Mendoza H, Perlin MH, Schirawski J. Mitochondrial Inheritance in Phytopathogenic Fungi—Everything Is Known, or Is It? International Journal of Molecular Sciences. 2020; 21(11):3883. https://doi.org/10.3390/ijms21113883
Chicago/Turabian StyleMendoza, Hector, Michael H. Perlin, and Jan Schirawski. 2020. "Mitochondrial Inheritance in Phytopathogenic Fungi—Everything Is Known, or Is It?" International Journal of Molecular Sciences 21, no. 11: 3883. https://doi.org/10.3390/ijms21113883
APA StyleMendoza, H., Perlin, M. H., & Schirawski, J. (2020). Mitochondrial Inheritance in Phytopathogenic Fungi—Everything Is Known, or Is It? International Journal of Molecular Sciences, 21(11), 3883. https://doi.org/10.3390/ijms21113883







