Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration
Abstract
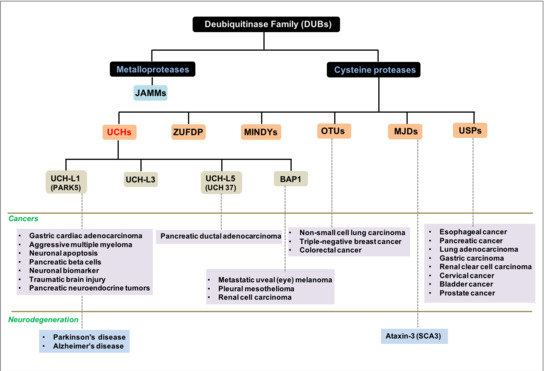
:1. Background
2. Results and Discussion
3. Conclusions
4. Material and Methods
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| USPs | ubiquitin-specific proteases |
| OTU | ovarian tumor proteases |
| MJDs | Machado–Josephin domain proteases |
| MINDYs | (motif interacting with Ub-containing novel DUB family) |
| ZUFDP | Zn-finger and UFSP domain protein |
| UCHs | ubiquitin C-terminal hydrolases |
| JAMM | Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) |
| SCA3 | Spinocerebellar ataxia type 3 |
References
- Poondla, N.; Chandrasekaran, A.P.; Kim, K.S.; Ramakrishna, S. Deubiquitinating enzymes as cancer biomarkers: New therapeutic opportunities? BMB Rep. 2019, 52, 181–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, C.; Sharma, A.; Peters, S.; Aretz, S.; Biswas, A.; Holz, F.G.; Loeffler, K.U.; Herwig-Carl, M.C. Basal cell carcinomas developing independently from BAP1-tumor predisposition syndrome in a patient with bilateral uveal melanoma: Diagnostic challenges to identify patients with BAP1-TPDS. Genes Chromosomes Cancer 2019, 58, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fu, D.; Shen, X.Z. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim. Biophys. Acta 2010, 1806, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Herwig-Carl, M.C.; Sharma, A.; Holler, T.; Holz, F.G.; Schlitter, A.M.; Loeffler, K.U. Spatial intratumor heterogeneity in uveal melanoma: Tumor cell subtypes with a presumed invasive potential exhibit a particular epigenetic staining reaction. Exp. Eye Res. 2019, 182, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Liu, H.; Tobar-Tosse, F.; Noll, A.; Dakal, T.C.; Li, H.; Holz, F.G.; Loeffler, K.; Herwig-Carl, M.C. Genome organization in proximity to the BAP1 locus appears to play a pivotal role in a variety of cancers. Cancer Sci. 2020, 111, 1385–1391. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Biswas, A.; Liu, H.; Sen, S.; Paruchuri, A.; Katsonis, P.; Lichtarge, O.; Chand Dakal, T.; Maulik, U.; Gromiha, M.M.; et al. Mutational Landscape of the BAP1 Locus Reveals an Intrinsic Control to Regulate the miRNA Network and the Binding of Protein Complexes in Uveal Melanoma. Cancers 2019, 11, 1600. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, K.D.; Lee, K.M.; Deshpande, S.; Duerksen-Hughes, P.; Boss, J.M.; Pohl, J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 1989, 246, 670–673. [Google Scholar] [CrossRef]
- Evert, B.O.; Araujo, J.; Vieira-Saecker, A.M.; de Vos, R.A.; Harendza, S.; Klockgether, T.; Wullner, U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J. Neurosci. 2006, 26, 11474–11486. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Ning, G.; Zhu, S.; Ma, X.; Liu, X.; Liu, C.; Huang, M.; Schmitt, I.; Wullner, U.; et al. The Machado-Joseph Disease Deubiquitinase Ataxin-3 Regulates the Stability and Apoptotic Function of p53. PLoS Biol. 2016, 14, e2000733. [Google Scholar] [CrossRef] [Green Version]
- Durcan, T.M.; Fon, E.A. Ataxin-3 and its e3 partners: Implications for machado-joseph disease. Front. Neurol. 2013, 4, 46. [Google Scholar] [CrossRef] [Green Version]
- Houck, A.L.; Seddighi, S.; Driver, J.A. At the Crossroads Between Neurodegeneration and Cancer: A Review of Overlapping Biology and Its Implications. Curr. Aging Sci. 2018, 11, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, K.; Boullosa, C.; Tabares-Seisdedos, R.; Baudot, A.; Valencia, A. Molecular evidence for the inverse comorbidity between central nervous system disorders and cancers detected by transcriptomic meta-analyses. PLoS Genet. 2014, 10, e1004173. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Meray, R.K.; Grammatopoulos, T.N.; Fredenburg, R.A.; Cookson, M.R.; Liu, Y.; Logan, T.; Lansbury, P.T., Jr. Membrane-associated farnesylated UCH-L1 promotes alpha-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2009, 106, 4635–4640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroy, E.; Boyer, R.; Auburger, G.; Leube, B.; Ulm, G.; Mezey, E.; Harta, G.; Brownstein, M.J.; Jonnalagada, S.; Chernova, T.; et al. The ubiquitin pathway in Parkinson’s disease. Nature 1998, 395, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Bilguvar, K.; Tyagi, N.K.; Ozkara, C.; Tuysuz, B.; Bakircioglu, M.; Choi, M.; Delil, S.; Caglayan, A.O.; Baranoski, J.F.; Erturk, O.; et al. Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 3489–3494. [Google Scholar] [CrossRef] [Green Version]
- Healy, D.G.; Abou-Sleiman, P.M.; Casas, J.P.; Ahmadi, K.R.; Lynch, T.; Gandhi, S.; Muqit, M.M.; Foltynie, T.; Barker, R.; Bhatia, K.P.; et al. UCHL-1 is not a Parkinson’s disease susceptibility gene. Ann. Neurol. 2006, 59, 627–633. [Google Scholar] [CrossRef]
- Zhang, M.; Cai, F.; Zhang, S.; Song, W. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) delays Alzheimer’s progression in vivo. Sci. Rep. 2014, 4, 7298. [Google Scholar] [CrossRef] [Green Version]
- Bonfanti, L.; Candeo, P.; Piccinini, M.; Carmignoto, G.; Comelli, M.C.; Ghidella, S.; Bruno, R.; Gobetto, A.; Merighi, A. Distribution of protein gene product 9.5 (PGP 9.5) in the vertebrate retina: Evidence that immunoreactivity is restricted to mammalian horizontal and ganglion cells. J. Comp. Neurol. 1992, 322, 35–44. [Google Scholar] [CrossRef]
- Nakajima, J.; Mekada, A.; Nakamura, J.; Nishida, Y.; Tokunaga, Y. Expression of protein gene product 9.5 in the anterior lens epithelial cells of atopic cataracts. J. Cataract Refract. Surg. 2002, 28, 2035–2039. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, M.; Ma, Y.; Luo, Q.; Liu, J.; Wang, J.; Yuan, X.; Sang, J.; Huang, C. UCHL1 acts as a colorectal cancer oncogene via activation of the beta-catenin/TCF pathway through its deubiquitinating activity. Int. J. Mol. Med. 2012, 30, 430–436. [Google Scholar] [CrossRef]
- Hurst-Kennedy, J.; Chin, L.S.; Li, L. Ubiquitin C-terminal hydrolase l1 in tumorigenesis. Biochem. Res. Int. 2012, 2012, 123706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Jamil, M.A.; Nuesgen, N.; Dauksa, A.; Gulbinas, A.; Schulz, W.A.; Oldenburg, J.; El-Maarri, O. Detailed methylation map of LINE-1 5’-promoter region reveals hypomethylated CpG hotspots associated with tumor tissue specificity. Mol. Genet. Genomic Med. 2019, 7, e601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamil, M.A.; Sharma, A.; Nuesgen, N.; Pezeshkpoor, B.; Heimbach, A.; Pavlova, A.; Oldenburg, J.; El-Maarri, O. F8 Inversions at Xq28 Causing Hemophilia A Are Associated With Specific Methylation Changes: Implication for Molecular Epigenetic Diagnosis. Front. Genet. 2019, 10, 508. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.; Lihua, S.; Zhe, Z.; Li, S.; Yoselin, P.; Michelle, P.; Sullivan Kathleen, E. Transposable element dysregulation in systemic lupus erythematosus and regulation by histone conformation and Hsp90. Clin. Immunol. 2018, 197, 6–18. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, A.; Liu, H.; Tobar-Tosse, F.; Chand Dakal, T.; Ludwig, M.; Holz, F.G.; Loeffler, K.U.; Wüllner, U.; Herwig-Carl, M.C. Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 3910. https://doi.org/10.3390/ijms21113910
Sharma A, Liu H, Tobar-Tosse F, Chand Dakal T, Ludwig M, Holz FG, Loeffler KU, Wüllner U, Herwig-Carl MC. Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration. International Journal of Molecular Sciences. 2020; 21(11):3910. https://doi.org/10.3390/ijms21113910
Chicago/Turabian StyleSharma, Amit, Hongde Liu, Fabian Tobar-Tosse, Tikam Chand Dakal, Michael Ludwig, Frank G. Holz, Karin U. Loeffler, Ullrich Wüllner, and Martina C. Herwig-Carl. 2020. "Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration" International Journal of Molecular Sciences 21, no. 11: 3910. https://doi.org/10.3390/ijms21113910
APA StyleSharma, A., Liu, H., Tobar-Tosse, F., Chand Dakal, T., Ludwig, M., Holz, F. G., Loeffler, K. U., Wüllner, U., & Herwig-Carl, M. C. (2020). Ubiquitin Carboxyl-Terminal Hydrolases (UCHs): Potential Mediators for Cancer and Neurodegeneration. International Journal of Molecular Sciences, 21(11), 3910. https://doi.org/10.3390/ijms21113910