DFNA5 (GSDME) c.991-15_991-13delTTC: Founder Mutation or Mutational Hotspot?
Abstract
1. Introduction
2. Results
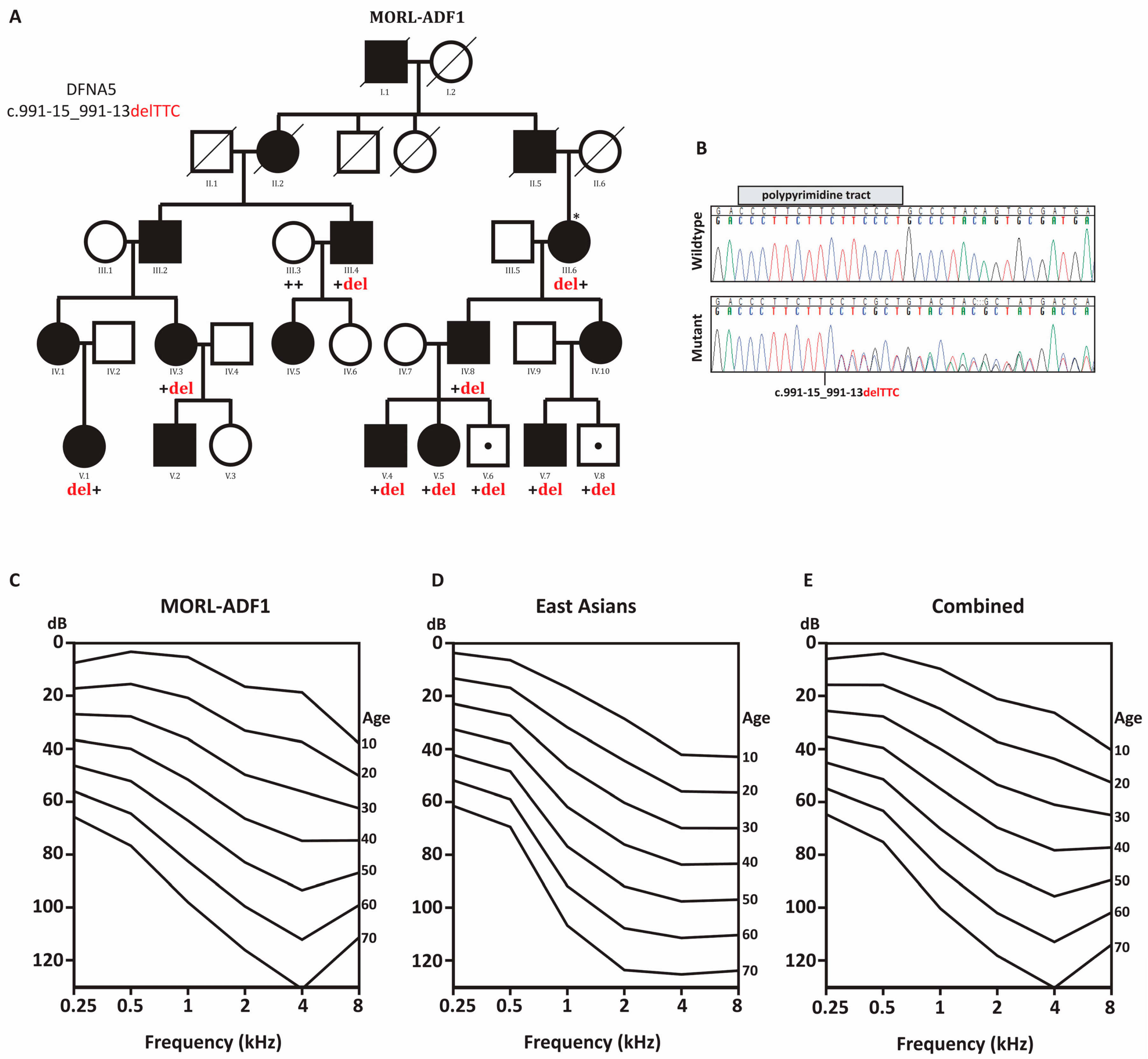
2.1. Clinical Presentation and Audiometric Analysis
2.2. OtoSCOPE®, Segregation Analysis, and Determining Ethnicity
2.3. Haplotype Analysis
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Audiometric Profiling
4.3. OtoSCOPE
4.4. Segregation Analysis
4.5. Ethnicity-Informative Markers
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Park, J.E.; Cartegni, L. In Vitro Modulation of Endogenous Alternative Splicing Using Splice-Switching Antisense Oligonucleotides. Methods Mol. Biol. 2017, 1648, 39–52. [Google Scholar] [CrossRef]
- Hentze, M.W.; Kulozik, A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 1999, 96, 307–310. [Google Scholar] [CrossRef]
- He, F.; Jacobson, A. Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story. Annu. Rev. Genet. 2015, 49, 339–366. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef]
- Cooper, T.A.; Wan, L.; Dreyfuss, G. RNA and Disease. Cell 2009, 136, 777–793. [Google Scholar] [CrossRef]
- Van Laer, L.; Huizing, E.H.; Verstreken, M.; van Zuijlen, D.; Wauters, J.G.; Bossuyt, P.J.; Van de Heyning, P.; McGuirt, W.T.; Smith, R.J.; Willems, P.J.; et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5. Nat. Genet. 1998, 20, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Booth, K.T.; Azaiez, H.; Kahrizi, K.; Wang, D.; Zhang, Y.; Frees, K.; Nishimura, C.; Najmabadi, H.; Smith, R.J. Exonic mutations and exon skipping: Lessons learned from DFNA5. Hum. Mutat. 2018, 39, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Nishio, A.; Noguchi, Y.; Sato, T.; Naruse, T.K.; Kimura, A.; Takagi, A.; Kitamura, K. A DFNA5 mutation identified in japanese families with autosomal dominant hereditary hearing loss. Ann. Hum. Genet. 2014. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-J.; Cho, H.-J.; Baek, J.-I.; Ben-Yosef, T.; Kwon, T.-J.; Griffith, A.J.; Kim, U.-K. Evidence for a founder mutation causing DFNA5 hearing loss in East Asians. J. Hum. Genet. 2010, 55, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Li-Yang, M.-N.N.; Shen, X.-F.F.; Wei, Q.-J.J.; Yao, J.; Lu, Y.-J.J.; Cao, X.; Xing, G.-Q.Q. IVS8+1 DelG, a Novel Splice Site Mutation Causing DFNA5 Deafness in a Chinese Family. Chin. Med. J. (Engl.) 2015, 128, 2510–2515. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Chen, D.; Wang, X.; Wu, H.; Yang, T. A novel splice site mutation in DFNA5 causes late-onset progressive non-syndromic hearing loss in a Chinese family. Int. J. Pediatr. Otorhinolaryngol. 2014, 78, 1265–1268. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Han, D.Y.; Dai, P.; Sun, H.J.; Tao, R.; Sun, Q.; Yan, D.; Qin, W.; Wang, H.Y.; Ouyang, X.M.; et al. A novel DFNA5 mutation, IVS8+4 A>G, in the splice donor site of intron 8 causes late-onset non-syndromic hearing loss in a Chinese family. Clin. Genet. 2007, 72, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Gregan, J.; Van Laer, L.; Lieto, L.D.; Van Camp, G.; Kearsey, S.E. A yeast model for the study of human DFNA5, a gene mutated in nonsyndromic hearing impairment. Biochim. Biophys. Acta Mol. Basis Dis. 2003, 1638, 179–186. [Google Scholar] [CrossRef][Green Version]
- Rogers, C.; Fernandes-Alnemri, T.; Mayes, L.; Alnemri, D.; Cingolani, G.; Alnemri, E.S. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun. 2017, 8, 14128. [Google Scholar] [CrossRef]
- Yu, C.; Meng, X.; Zhang, S.; Zhao, G.; Hu, L.; Kong, X. A 3-nucleotide deletion in the polypyrimidine tract of intron 7 of the DFNA5 gene causes nonsyndromic hearing impairment in a Chinese family. Genomics 2003. [Google Scholar] [CrossRef]
- Wang, H.; Guan, J.; Guan, L.; Yang, J.; Wu, K.; Lin, Q.; Xiong, W.; Lan, L.; Zhao, C.; Xie, L.; et al. Further evidence for “gain-of-function” mechanism of DFNA5 related hearing loss. Sci. Rep. 2018, 8, 8424. [Google Scholar] [CrossRef]
- Van Camp, G.; Coucke, P.J.; Akita, J.; Fransen, E.; Abe, S.; De Leenheer, E.M.R.; Huygen, P.L.M.; Cremers, C.W.R.J.; Usami, S.-I. A mutational hot spot in theKCNQ4 gene responsible for autosomal dominant hearing impairment. Hum. Mutat. 2002, 20, 15–19. [Google Scholar] [CrossRef]
- Wang, L.; Yan, D.; Qin, L.; Li, T.; Liu, H.; Li, W.; Mittal, R.; Yong, F.; Chapagain, P.; Liao, S.; et al. Amino acid 118 in the deafness causing (DFNA20/26) ACTG1 gene is a mutational hot spot. Gene Rep. 2018, 11, 264–269. [Google Scholar] [CrossRef]
- Choi, H.J.; Lee, J.S.; Yu, S.; Cha, D.H.; Gee, H.Y.; Choi, J.Y.; Lee, J.D.; Jung, J. Whole-exome sequencing identified a missense mutation in WFS1 causing low-frequency hearing loss: A case report. BMC Med. Genet. 2017, 18, 151. [Google Scholar] [CrossRef]
- Moteki, H.; Nishio, S.; Hashimoto, S.; Takumi, Y.; Iwasaki, S.; Takeichi, N.; Fukuda, S.; Usami, S. TECTA mutations in Japanese with mid-frequency hearing loss affected by zona pellucida domain protein secretion. J. Hum. Genet. 2012, 57, 587–592. [Google Scholar] [CrossRef]
- Taylor, K.R.; Booth, K.T.; Azaiez, H.; Sloan, C.M.; Kolbe, D.L.; Glanz, E.N.; Shearer, A.E.; DeLuca, A.P.; Anand, V.N.; Hildebrand, M.S.; et al. Audioprofile surfaces: The 21st century audiogram. Ann. Otol. Rhinol. Laryngol. 2016, 125, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Huygen, P.L.M.; Pennings, R.J.E.; Cremers, C.W.R.J. Characterizing and Distinguishing Progressive Phenotypes in Nonsyndromic Autosomal Dominant Hearing Impairment. Audiol. Med. 2003, 1, 37–46. [Google Scholar] [CrossRef]
- Daniel, W.; Kevin, B.; Azaiez Hela, S.R.J. A Comparative Analysis of Genetic Hearing Loss Phenotypes in European/American and Japanese Populations. Hum. Genet. 2020. In Press. [Google Scholar]
- Azaiez, H.; Decker, A.R.; Booth, K.T.; Simpson, A.C.; Shearer, A.E.; Huygen, P.L.M.; Bu, F.; Hildebrand, M.S.; Ranum, P.T.; Shibata, S.B.; et al. HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice. PLoS Genet. 2015, 11, e1005137. [Google Scholar] [CrossRef]
- Azaiez, H.; Booth, K.T.; Bu, F.; Huygen, P.; Shibata, S.B.; Shearer, A.E.; Kolbe, D.; Meyer, N.; Black-Ziegelbein, E.A.; Smith, R.J.H. TBC1D24 Mutation Causes Autosomal-Dominant Nonsyndromic Hearing Loss. Hum. Mutat. 2014, 35, 819–823. [Google Scholar] [CrossRef]
- Booth, K.T.; Azaiez, H.; Kahrizi, K.; Simpson, A.C.; Tollefson, W.T.A.; Sloan, C.M.; Meyer, N.C.; Babanejad, M.; Ardalani, F.; Arzhangi, S.; et al. PDZD7 and hearing loss: More than just a modifier. Am. J. Med. Genet. Part A 2015, 167, 2957–2965. [Google Scholar] [CrossRef]
- Booth, K.T.; Kahrizi, K.; Najmabadi, H.; Azaiez, H.; Smith, R.J. Old gene, new phenotype: Splice-altering variants in CEACAM16 cause recessive non-syndromic hearing impairment. J. Med. Genet. 2018, 55, 555–560. [Google Scholar] [CrossRef]
- Azaiez, H.; Booth, K.T.; Ephraim, S.S.; Crone, B.; Black-Ziegelbein, E.A.; Marini, R.J.; Shearer, A.E.; Sloan-Heggen, C.M.; Kolbe, D.; Casavant, T.; et al. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am. J. Hum. Genet. 2018, 103, 484–497. [Google Scholar] [CrossRef]
- Nord, A.S.; Lee, M.; King, M.-C.; Walsh, T. Accurate and exact CNV identification from targeted high-throughput sequence data. Bmc Genom. 2011, 12, 184. [Google Scholar] [CrossRef]
- Kidd, K.K.; Speed, W.C.; Pakstis, A.J.; Furtado, M.R.; Fang, R.; Madbouly, A.; Maiers, M.; Middha, M.; Friedlaender, F.R.; Kidd, J.R. Progress toward an efficient panel of SNPs for ancestry inference. Forensic. Sci. Int. Genet. 2014, 10, 23–32. [Google Scholar] [CrossRef]
| 0.250 kHz | 0.500 kHz | 1 kHz | 2 kHz | 4 kHz | 8 kHz | |
|---|---|---|---|---|---|---|
| MORL-ADF1 | 0.9685 | 1.222 | 1.543 | 1.659 | 1.869 | 1.221 |
| E. Asians | 0.9629 | 1.050 | 1.499 | 1.584 | 1.385 | 1.349 |
| Difference | 0.0056 | 0.172 | 0.044 | 0.075 | 0.484 | 0.128 |
| p-value | 0.186 | 0.391 | 0.691 | 0.767 | 0.194 | 0.829 |
| Combined | 0.977 | 1.186 | 1.508 | 1.618 | 1.733 | 1.227 |
| SNP | Loc | ADF1-III.6 | Korean [7] | Korean [9] | Chinese [9] | Japanese [8] | MAF European (NF) | MAF E. Asian | MAF Global | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17149912 (T|C) | Ex 9 | T|T | T|C | T | C | C | C | 15.69% | 27.68% | 19.78% |
| rs2240005 (G|A) | In 8 | G|A | G|G | G | G | G | G | 22.87% | 24.33% | 30.78% |
| rs66851582 (C|T) | In 8 | C|T | C|C | - | - | - | - | 14.17% | 0.06% | 10.88% |
| rs2074142 (C|T) | In 8 | C|C | C|T | T | T | T | T | 25.27% | 67.16% | 34.30% |
| rs727505273 (GAA|Del) | In 7 | GAA|Del | GAA|Del | Del | Del | Del | Del | 0.00% | 0.00% | 0.00% |
| rs17209408 (C|T) | In 7 | C|T | C|C | C | C | C | C | 2.80% | 0.01% | 1.77% |
| rs141596134 (C|T) | Ex 7 | C|C | C|T | - | - | - | - | 0.00% | 0.10% | 0.01% |
| rs2721809 (G|A) | In 6 | G|G | A|A | - | - | - | - | 42.28% | 99.14% | 56.34% |
| rs35529766 (C|Del) | In 6 | C|C | C|Del | Del | Del | Del | Del | 0.00% | 0.00% | 0.00% |
| rs10601416; 35521389 (TA|Del) | In 4 | TA|TA | Del|Del | - | - | - | - | 49.08% | 99.23% | 60.79% |
| rs876308 (G|A) | In 4 | G|A | G|G | - | - | - | - | 43.60% | 99.17% | 56.98% |
| rs876307 (G|T) | In 4 | G|G | T|T | - | - | - | - | 42.46% | 99.12% | 57.01% |
| rs754553 (C|T) | In 3 | C|C | C|T | T | T | T | T | 15.04% | 45.96% | 19.80% |
| rs2023793 (G|A) | In 3 | G|G | A|A | A | A | - | - | 43.50% | 99.23% | 56.92% |
| rs2521768 (C|T) | In 2 | C|C | T|T | T | T | T | T | 46.55% | 79.95% | 58.43% |
| rs150598245 (wt|insGT) | In 2 | wt|GT | wt|GT | - | - | - | - | 1.68% | 0.98% | 1.30% |
| rs2521770 (C|T) | In 2 | C|T | T|T | - | - | - | - | 51.88% | 99.87% | 65.31% |
| rs768391255 (wt|Ins) | In 2 | wt|Ins | -|- | - | - | - | - | 16.41% | 21.58% | 14.58% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Booth, K.T.; Azaiez, H.; Smith, R.J.H. DFNA5 (GSDME) c.991-15_991-13delTTC: Founder Mutation or Mutational Hotspot? Int. J. Mol. Sci. 2020, 21, 3951. https://doi.org/10.3390/ijms21113951
Booth KT, Azaiez H, Smith RJH. DFNA5 (GSDME) c.991-15_991-13delTTC: Founder Mutation or Mutational Hotspot? International Journal of Molecular Sciences. 2020; 21(11):3951. https://doi.org/10.3390/ijms21113951
Chicago/Turabian StyleBooth, Kevin T., Hela Azaiez, and Richard J. H. Smith. 2020. "DFNA5 (GSDME) c.991-15_991-13delTTC: Founder Mutation or Mutational Hotspot?" International Journal of Molecular Sciences 21, no. 11: 3951. https://doi.org/10.3390/ijms21113951
APA StyleBooth, K. T., Azaiez, H., & Smith, R. J. H. (2020). DFNA5 (GSDME) c.991-15_991-13delTTC: Founder Mutation or Mutational Hotspot? International Journal of Molecular Sciences, 21(11), 3951. https://doi.org/10.3390/ijms21113951





