Heterologous Expression of Nitrate Assimilation Related-Protein DsNAR2.1/NRT3.1 Affects Uptake of Nitrate and Ammonium in Nitrogen-Starved Arabidopsis
Abstract
1. Introduction
2. Results
2.1. Expression Analysis of DsNRT3.1
2.2. Effect of Heterologous Expression of DsNRT3.1 on Arabidopsis Seedings Growth
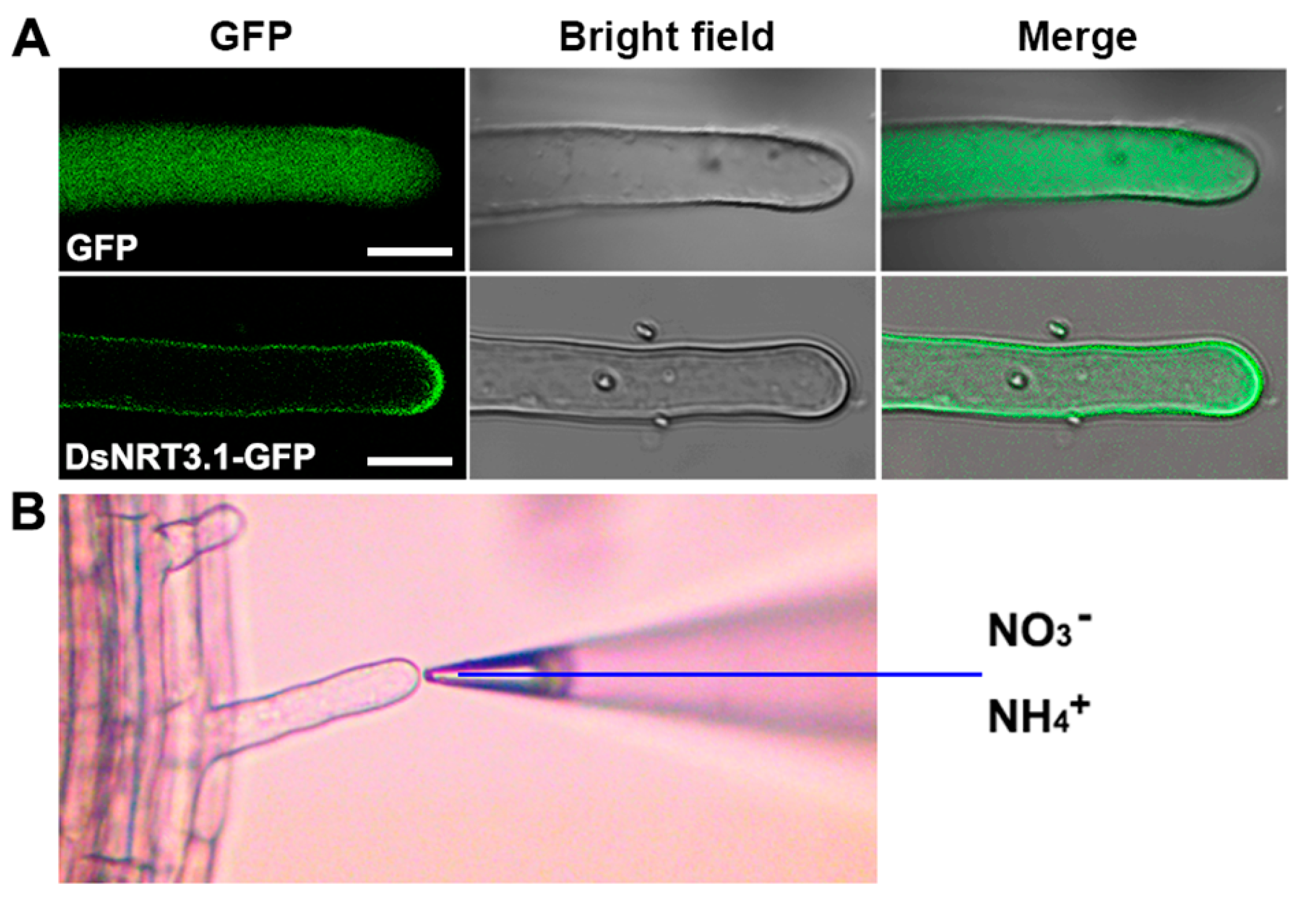
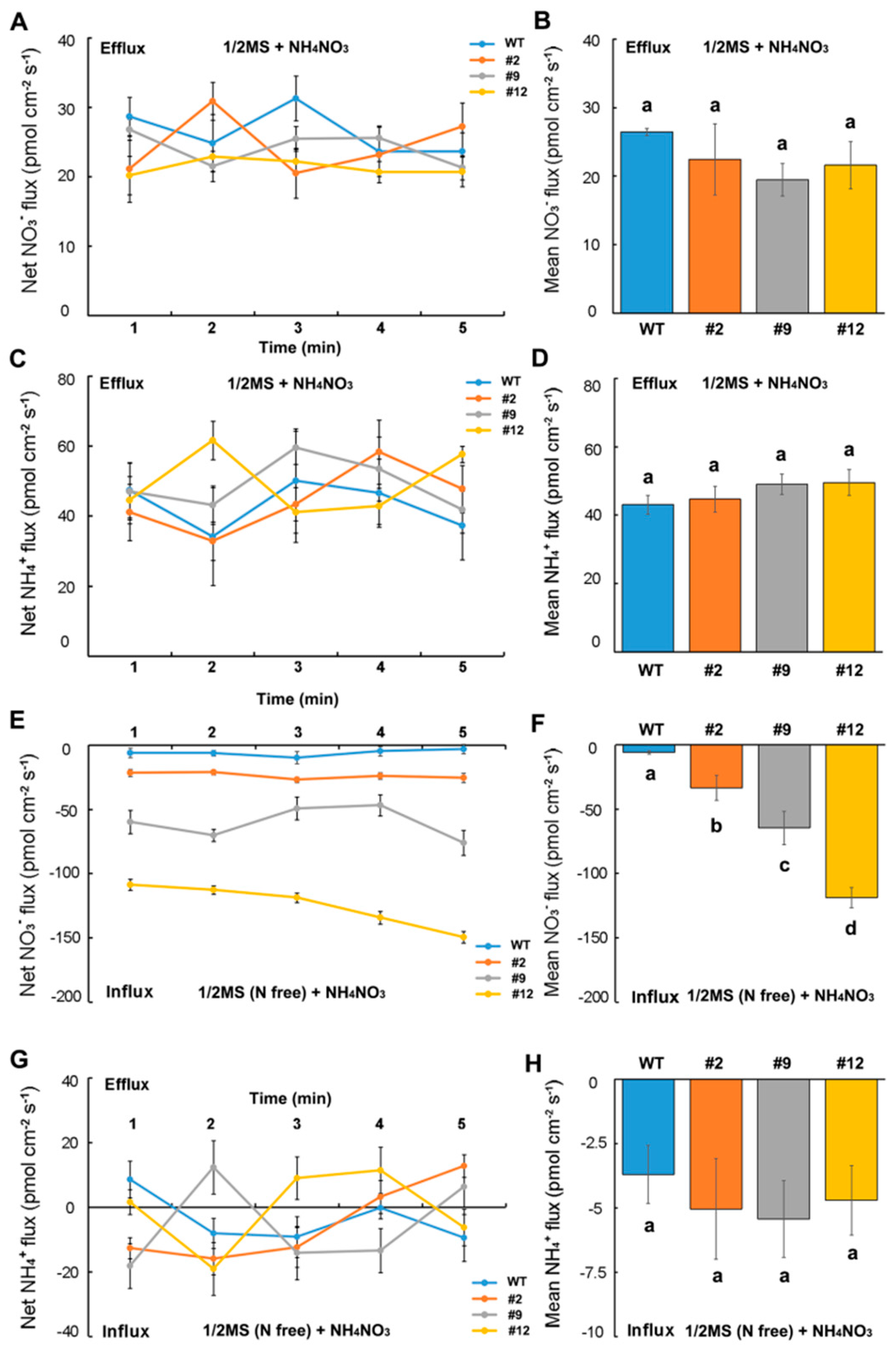
2.3. Effect of Heterologous Expression of DsNRT3.1 on the NO3− and NH4+ Fluxes in Arabidopsis Root Hairs
2.4. Effect of Heterologous Expression of DsNRT3.1 on Arabidopsis NRT2 Genes Expression
3. Discussion
4. Materials and Methods
4.1. Sequence Analysis of DsNRT3.1
4.2. Plant Materials and Growth Conditions
4.3. RNA Preparation and Expression Analysis
4.4. Vector Construction and Plant Transformation
4.5. Subcellular Localization of DsNRT3.1
4.6. Phenotypic Analysis of Transformed Arabidopsis
4.7. NMT Measurement of Net NO3− and NH4+ Fluxes in Arabidopsis Root Hairs
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Xu, G.H.; Fan, X.R.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, T.; Sakakibara, H. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 2017, 68, 2501–2512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.X.; Li, Y.; Yang, X.X.; Li, H.J.; Shen, Q.R.; Guo, S.W. Ammonium nutrition increases water absorption in rice seedlings (Oryza sativa L.) under water stress. Plant Soil 2010, 331, 193–201. [Google Scholar] [CrossRef]
- Ruan, L.; Wei, K.; Wang, L.Y.; Cheng, H.; Zhang, F.; Wu, L.Y.; Bai, P.X.; Zhang, C.C. Characteristics of NH4+ and NO3− fluxes in tea (Camellia sinensis) roots measured by scanning ion-selective electrode technique. Sci. Rep. 2016, 6, 38370. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Naz, M.; Fan, X.; Xuan, W.; Miller, A.J.; Xu, G. Plant nitrate transporters: From gene function to application. J. Exp. Bot. 2017, 68, 2463–2475. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutierrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef]
- Leran, S.; Varala, K.; Boyer, J.C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Von Wittgenstein, N.J.; Le, C.H.; Hawkins, B.J.; Ehlting, J. Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol. Biol. 2014, 14, 11. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Hsu, P.K.; Tsay, Y.F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef]
- Miller, A.J.; Fan, X.R.; Orsel, M.; Smith, S.J.; Wells, D.M. Nitrate transport and signalling. J. Exp. Bot. 2007, 58, 2297–2306. [Google Scholar] [CrossRef]
- Feng, H.; Yan, M.; Fan, X.; Li, B.; Shen, Q.; Miller, A.J.; Xu, G. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 2011, 62, 2319–2332. [Google Scholar] [CrossRef]
- Quesada, A.; Galvan, A.; Fernandez, E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. Plant J. 1994, 5, 407–419. [Google Scholar] [CrossRef]
- Yong, Z.; Kotur, Z.; Glass, A.D. Characterization of an intact two-component high-affinity nitrate transporter from Arabidopsis roots. Plant J. 2010, 63, 739–748. [Google Scholar] [CrossRef]
- Kotur, Z.; Mackenzie, N.; Ramesh, S.; Tyerman, S.D.; Kaiser, B.N.; Glass, A.D. Nitrate transport capacity of the Arabidopsis thaliana NRT2 family members and their interactions with AtNAR2.1. New Phytol. 2012, 194, 724–731. [Google Scholar] [CrossRef]
- Okamoto, M.; Kumar, A.; Li, W.; Wang, Y.; Siddiqi, M.Y.; Crawford, N.M.; Glass, A.D. High-affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2-like gene AtNRT3.1. Plant Physiol. 2006, 140, 1036–1046. [Google Scholar] [CrossRef]
- Kotur, Z.; Glass, A.D. A 150 kDa plasma membrane complex of AtNRT2.5 and AtNAR2.1 is the major contributor to constitutive high-affinity nitrate influx in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 1490–1502. [Google Scholar] [CrossRef]
- Yan, M.; Fan, X.; Feng, H.; Miller, A.J.; Shen, Q.; Xu, G. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 2011, 34, 1360–1372. [Google Scholar] [CrossRef]
- Liu, X.; Huang, D.; Tao, J.; Miller, A.J.; Fan, X.; Xu, G. Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol. 2014, 204, 74–80. [Google Scholar] [CrossRef]
- Zhou, A.; Ma, H.; Liu, E.; Jiang, T.; Feng, S.; Gong, S.; Wang, J. Transcriptome sequencing of Dianthus spiculifolius and analysis of the genes involved in responses to combined cold and drought stress. Int. J. Mol. Sci. 2017, 18, 849. [Google Scholar] [CrossRef]
- Bohme, K.; Li, Y.; Charlot, F.; Grierson, C.; Marrocco, K.; Okada, K.; Laloue, M.; Nogue, F. The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 2004, 40, 686–698. [Google Scholar] [CrossRef]
- Tong, Y.; Zhou, J.J.; Li, Z.; Miller, A.J. A two-component high-affinity nitrate uptake system in barley. Plant J. 2005, 41, 442–450. [Google Scholar] [CrossRef]
- Gu, C.; Zhang, X.; Jiang, J.; Guan, Z.; Zhao, S.; Fang, W.; Liao, Y.; Chen, S.; Chen, F. Chrysanthemum CmNAR2 interacts with CmNRT2 in the control of nitrate uptake. Sci. Rep. 2014, 4, 5833. [Google Scholar] [CrossRef]
- Li, H.; Yu, M.; Du, X.Q.; Wang, Z.F.; Wu, W.H.; Quintero, F.J.; Jin, X.H.; Li, H.D.; Wang, Y. NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell 2017, 29, 2016–2026. [Google Scholar] [CrossRef]
- Lezhneva, L.; Kiba, T.; Feria-Bourrellier, A.B.; Lafouge, F.; Boutet-Mercey, S.; Zoufan, P.; Sakakibara, H.; Daniel-Vedele, F.; Krapp, A. The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J. 2014, 80, 230–241. [Google Scholar] [CrossRef]
- Jian, S.; Liao, Q.; Song, H.; Liu, Q.; Lepo, J.E.; Guan, C.; Zhang, J.; Ismail, A.M.; Zhang, Z. NRT1.1-related NH4+ toxicity is associated with a disturbed balance between NH4+ uptake and assimilation. Plant Physiol. 2018, 178, 1473–1488. [Google Scholar] [CrossRef]
- Zhou, A.; Bu, Y.; Takano, T.; Zhang, X.; Liu, S. Conserved V-ATPase c subunit plays a role in plant growth by influencing V-ATPase-dependent endosomal trafficking. Plant Biotechnol. J. 2016, 14, 271–283. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Bai, L.; Ma, X.; Zhang, G.; Song, S.; Zhou, Y.; Gao, L.; Miao, Y.; Song, C.P. A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. Plant Cell 2014, 26, 1497–1511. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, H.; Zhao, J.; Feng, S.; Qiao, K.; Gong, S.; Wang, J.; Zhou, A. Heterologous Expression of Nitrate Assimilation Related-Protein DsNAR2.1/NRT3.1 Affects Uptake of Nitrate and Ammonium in Nitrogen-Starved Arabidopsis. Int. J. Mol. Sci. 2020, 21, 4027. https://doi.org/10.3390/ijms21114027
Ma H, Zhao J, Feng S, Qiao K, Gong S, Wang J, Zhou A. Heterologous Expression of Nitrate Assimilation Related-Protein DsNAR2.1/NRT3.1 Affects Uptake of Nitrate and Ammonium in Nitrogen-Starved Arabidopsis. International Journal of Molecular Sciences. 2020; 21(11):4027. https://doi.org/10.3390/ijms21114027
Chicago/Turabian StyleMa, Hongping, Junchao Zhao, Shuang Feng, Kun Qiao, Shufang Gong, Jingang Wang, and Aimin Zhou. 2020. "Heterologous Expression of Nitrate Assimilation Related-Protein DsNAR2.1/NRT3.1 Affects Uptake of Nitrate and Ammonium in Nitrogen-Starved Arabidopsis" International Journal of Molecular Sciences 21, no. 11: 4027. https://doi.org/10.3390/ijms21114027
APA StyleMa, H., Zhao, J., Feng, S., Qiao, K., Gong, S., Wang, J., & Zhou, A. (2020). Heterologous Expression of Nitrate Assimilation Related-Protein DsNAR2.1/NRT3.1 Affects Uptake of Nitrate and Ammonium in Nitrogen-Starved Arabidopsis. International Journal of Molecular Sciences, 21(11), 4027. https://doi.org/10.3390/ijms21114027




