Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance
Abstract
1. Introduction
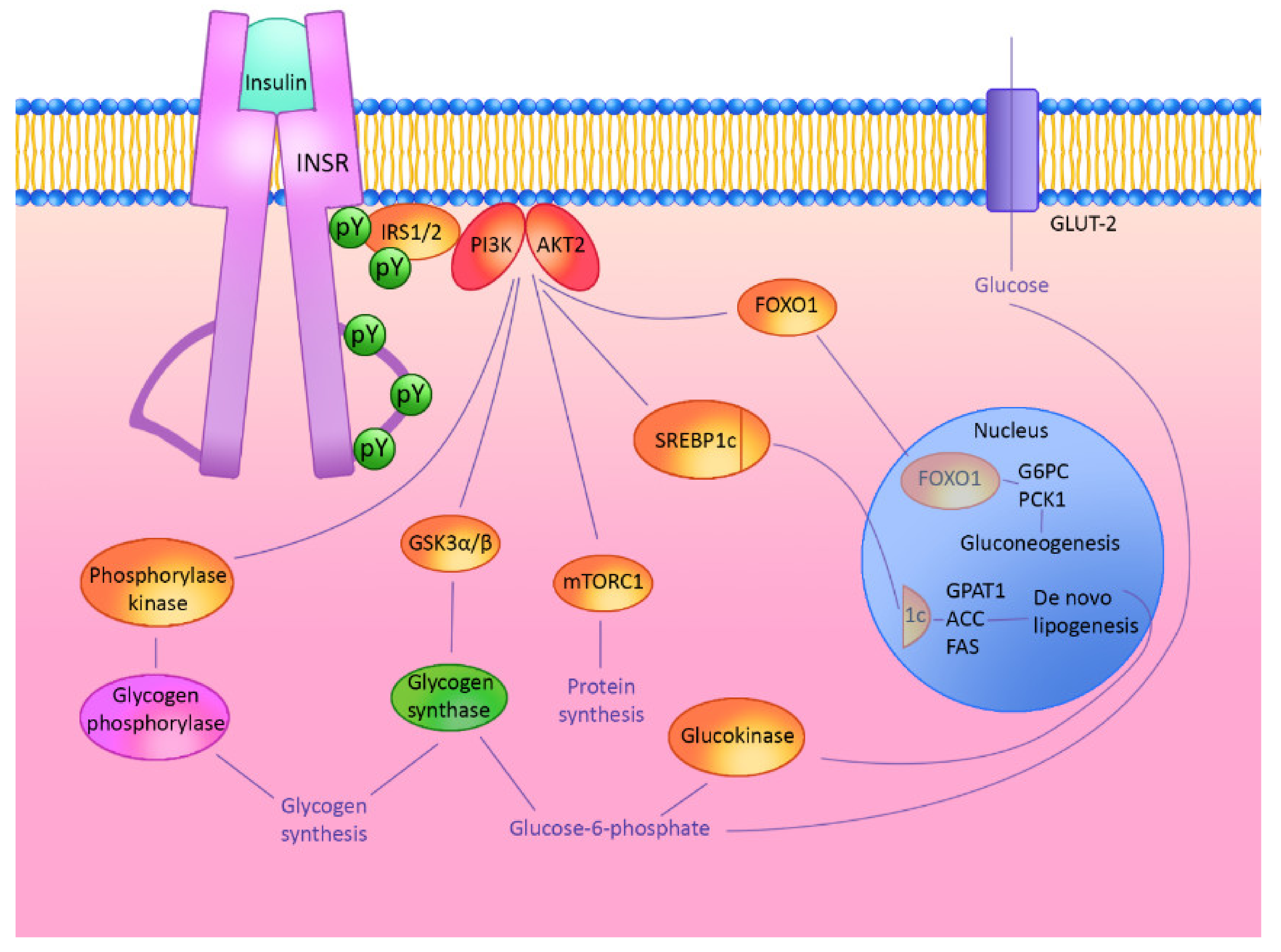
2. Hepatic Insulin Signaling Cascade
3. microRNAs and lncRNAs—Crucial Regulators of Cellular Pathways
3.1. miR-802
3.2. miR 499-5p
3.3. miR 122-5p
3.4. Long-Non-Coding RNA MALAT1(Metastasis Associated in Lung Adenocarcinoma Transcript 1)
3.5. Long-Non-Coding RNA MEG3
3.6. Long-Non-Coding RNA H19
4. Prognostic Potential
5. Summary and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MetS | Metabolic Syndrome |
| T2D | Type 2 Diabetes |
| NAFLD | Nonalcoholic Fatty Liver Disease |
| HCC | Hepatocellular Carcinoma |
| WHO | World Health Organization |
| FPG | Fasting Plasma Glucose |
| 2-h PH | 2-h Plasma Glucose |
| OGTT | Oral Glucose Tolerance Test |
| HbA1C | Hemoglobin A1c |
| FSIVGTT | Frequently-Sampled Intravenous Glucose Tolerance Test |
| HOMA-IR | Homeostatic Model Assessment of Insulin Resistance |
| HOMA2 | Homeostatic Model Assessment 2 |
| QUICKI | Quantitative Insulin Sensitivity Check Index |
| PCOS | Polycystic Ovary Syndrome |
| ncRNA | Non-Coding RNA |
| lncRNA | Long Non-Coding RNA |
| pre-miRNA | Precursor miRNA |
| miRNA | microRNA |
| (ce)RNA | Competing Endogenous RNA |
| Nts | Nucleotides |
| mRNA | Messenger RNA |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 |
| MEG3 | Maternally Expressed Gene 3 |
| H19 | H19 Imprinted Maternally Expressed Transcript |
| HFD-mice | High Fat Diet Mice |
| Lepr db/db | Homozygous for the Diabetes db Mutation of the Leptin Receptor |
| GDM | Gestational Diabetes Mellitus |
| INSR | Insulin Receptor |
| IRS | Insulin Receptor Substrate |
| IRS1 | Insulin Receptor Substrate 1 |
| IRS2 | Insulin Receptor Substrate 2 |
| Pi3K | Phosphatidylinositol 3-kinase |
| PIP3 | Prolactin Induced Protein 3 |
| PIP2 | Prolactin Induced Protein 2 |
| Akt | Protein Kinase B |
| PTEN | Phosphatase and Tensin Homolog |
| FOXO | Forkhead Box |
| FOXO1 | Forkhead Box O1 |
| GSK3 | Glycogen Synthase Kinase 3 |
| mTOR | Mechanistic Target of Rapamycin Kinase |
| G6PC | Glucose-6-Phosphatase Catalytic Subunit |
| PCK1 | Phosphoenolpyruvate Carboxykinase 1 |
| SREBP-1c | Sterol Regulatory Element-Binding Proteins |
| ROS | Reactive Oxygen Species |
| SOD | Superoxide Dismutase |
| CAT | Catalase |
| GSH-Px | Glutathione Peroxidase |
| Hnf1b | Hepatocyte Nuclear Factor 1-beta |
| PPARγ | Peroxisome Proliferator-Activated Receptor Gamma Coactivator |
| Myh7b | Beta-Myosin Heavy Chain |
| MMP-7 | Matrix Metallopeptidase 7 |
| SPF | Specific Pathogen-Free |
| HNF6 | Hepatocyte Nuclear Factor 6 |
| OC2 | One Cut Homeobox 2 |
| BMEL | Bipotent Murine Embryonic Liver Cells |
| FASN | Fatty Acid Synthase |
| IR | Insulin-Resistant |
| Acc1 | Acetyl-CoA Carboxylase |
| HNF-4α | Hepatocyte Nuclear Factor 4 α |
| NEAT2 | Noncoding Nuclear-Enriched Abundant Transcript 2 |
| HUVECs | Human Umbilical Vein Endothelial Cells |
| IL-6 | Interleukin-6 |
| TNF-α | Tumor Necrosis Factor α |
| SAA3 | Serum Amyloid A3 |
| JNK | c-Jun N-Terminal Kinase |
| DLK1-MEG3 | Imprinted Delta Like Non-Canonical Notch Ligand |
| GTL2 | Gene Trap Locus 2 |
| NSCLC | Non-Small Cell Lung Cancer |
| ATF4 | Activating Transcription Factor 4 |
| EGR2 | Early Growth Response 2 |
| PBMC | Peripheral Blood Mononuclear Cells |
| IGF2 | Insulin-Like Growth Factor 2 |
| IGN | Imprinted Gene Network |
| RNA-seq | RNA Sequencing |
| SCD1 | Stearoyl-CoA Desaturase |
| HDL-C | High-density Lipoprotein Cholesterol |
| eGRF | Estimated Glomerular Filtration Rate Test |
| ANIT | Alpha-Naphthylisothiocyanate |
| HBV | Hepatitis B Virus |
| AUC | Area Under Curve |
| ROC | Receiver Operating Characteristics |
References
- Internation Diabetes Federation. IDF Diabetes Atlas Ninth; International Diabetes Federation: Brussels, Belgium, 2019; ISBN 9782930229874. [Google Scholar]
- Ranasinghe, P.; Mathangasinghe, Y.; Jayawardena, R.; Hills, A.P.; Misra, A. Prevalence and trends of metabolic syndrome among adults in the Asia-pacific region: A systematic review. BMC Public Health 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.X.; Chaudhary, N.; Akinyemiju, T. Metabolic syndrome prevalence by race/ ethnicity and sex in the united states, national health and nutrition examination survey, 1988–2012. Prev. Chronic Dis. 2017, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Shulman, G.I. Mechanisms of insulin action and insulin resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef] [PubMed]
- Han, H.S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, 1–10. [Google Scholar] [CrossRef]
- Caputo, T.; Gilardi, F.; Desvergne, B. From chronic overnutrition to metaflammation and insulin resistance: Adipose tissue and liver contributions. FEBS Lett. 2017, 591, 3061–3088. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, R.; Xiong, Y.; Du, F.; Zhu, S. A vicious circle between insulin resistance and inflammation in nonalcoholic fatty liver disease. Lipids Health Dis. 2017, 16, 1–9. [Google Scholar] [CrossRef]
- Johnson, A.M.F.; Olefsky, J.M. The origins and drivers of insulin resistance. Cell 2013, 152, 673–684. [Google Scholar] [CrossRef]
- Lonardo, A.; Ballestri, S.; Marchesini, G.; Angulo, P.; Loria, P. Nonalcoholic fatty liver disease: A precursor of the metabolic syndrome. Dig. Liver Dis. 2015, 47, 181–190. [Google Scholar] [CrossRef]
- Dvorak, K.; Hainer, R.; Petrtyl, J.; Zeman, M.; Vareka, T.; Zak, A.; Sroubkova, R.; Svestka, T.; Vitek, L.; Bruha, R. The prevalence of nonalcoholic liver steatosis in patients with type 2 diabetes mellitus in the Czech Republic. Biomed. Pap. 2015, 159, 442–448. [Google Scholar] [CrossRef]
- Koehler, E.M.; Plompen, E.P.C.; Schouten, J.N.L.; Hansen, B.E.; Darwish Murad, S.; Taimr, P.; Leebeek, F.W.G.; Hofman, A.; Stricker, B.H.; Castera, L.; et al. Presence of diabetes mellitus and steatosis is associated with liver stiffness in a general population: The Rotterdam study. Hepatology 2016, 63, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Care, D. Classification and diagnosis of diabetes: Standards of medical care in diabetes 2019. Diabetes Care 2019, 42, S13–S28. [Google Scholar]
- Meijnikman, A.S.; De Block, C.E.M.; Dirinck, E.; Verrijken, A.; Mertens, I.; Corthouts, B.; Van Gaal, L.F. Not performing an OGTT results in significant underdiagnosis of (pre)diabetes in a high risk adult Caucasian population. In International Journal of Obesity; Nature Publishing Group, 2017; Volume 41, pp. 1615–1620. [Google Scholar]
- Karnchanasorn, R.; Huang, J.; Ou, H.Y.; Feng, W.; Chuang, L.M.; Chiu, K.C.; Samoa, R. Comparison of the Current Diagnostic Criterion of HbA1c with Fasting and 2-Hour Plasma Glucose Concentration. J. Diabetes Res. 2016, 2016, 6195494. [Google Scholar] [CrossRef] [PubMed]
- Placzkowska, S.; Pawlik-Sobecka, L.; Kokot, I.; Piwowar, A. Indirect insulin resistance detection: Current clinical trends and laboratory limitations. Biomed. Pap. 2019, 163, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Courtney, C.H.; Olefsky, J.M. Insulin resistance. In Mechanisms of Insulin Action: Medical Intelligence Unit; Springer New York: New York, NY, USA, 2007; pp. 185–209. ISBN 9780387722030. [Google Scholar]
- Muniyappa, R.; Madan, R. Assessing Insulin Sensitivity and Resistance in Humans; MDText.com, Inc.: Bethesda, MD, USA, 2000. [Google Scholar]
- Tosi, F.; Bonora, E.; Moghetti, P. Insulin resistance in a large cohort of women with polycystic ovary syndrome: A comparison between euglycaemic-hyperinsulinaemic clamp and surrogate indexes. Hum. Reprod. 2017, 32, 2515–2521. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Leung, S.W. Identification of microRNA biomarkers in type 2 diabetes: A meta-analysis of controlled profiling studies. Diabetologia 2015, 58, 900–911. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, J.; Tang, X.; Li, Y.; Xia, P.; Gao, X. Berberine ameliorates nonalcoholic fatty liver disease by a global modulation of hepatic mRNA and lncRNA expression profiles. J. Transl. Med. 2015, 13, 24. [Google Scholar] [CrossRef]
- Kornfeld, J.W.; Baitzel, C.; Könner, A.C.; Nicholls, H.T.; Vogt, M.C.; Herrmanns, K.; Scheja, L.; Haumaitre, C.; Wolf, A.M.; Knippschild, U.; et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 2013, 494, 111–115. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, H.; Wang, F.; Ye, M.; Zhu, H.; Bu, S. Long non-coding RNA MALAT1 expression in patients with gestational diabetes mellitus. Int. J. Gynecol. Obstet. 2018, 140, 164–169. [Google Scholar] [CrossRef]
- Higuchi, C.; Nakatsuka, A.; Eguchi, J.; Teshigawara, S.; Kanzaki, M.; Katayama, A.; Yamaguchi, S.; Takahashi, N.; Murakami, K.; Ogawa, D.; et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015, 64, 489–497. [Google Scholar] [CrossRef]
- Yang, X.; Xing, H.; Liu, J.; Yang, L.; Ma, H.; Ma, H. MicroRNA-802 increases hepatic oxidative stress and induces insulin resistance in high-fat fed mice. Mol. Med. Rep. 2019, 20, 1230–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, N.; Pan, H.P.; Wang, Z.; Cao, Z.Y. MiR-499-5p contributes to hepatic insulin resistance by suppressing PTEN. Cell. Physiol. Biochem. 2015, 36, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Fluitt, M.B.; Kumari, N.; Nunlee-Bland, G.; Nekhai, S.; Gambhir, K.K. MiRNA-15a, miRNA-15b, and miRNA-499 are Reduced in Erythrocytes of Pre-Diabetic African-American Adults. Jacobs J. Diabetes Endocrinol. 2016, 2. [Google Scholar]
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 2008, 48, 1810–1820. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of MicroRNA-122 and Cpt1α and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Hou, X.; Liu, F.; Tao, H.; Zhang, Y.; Zhao, H.; Song, G. Regulation of insulin resistance by targeting the insulin-like growth factor 1 receptor with microRNA-122-5p in hepatic cells. Cell Biol. Int. 2019, 43, 553–564. [Google Scholar] [CrossRef]
- Wei, S.; Zhang, M.; Yu, Y.; Xue, H.; Lan, X.; Liu, S.; Hatch, G.; Chen, L. HNF-4α regulated miR-122 contributes to development of gluconeogenesis and lipid metabolism disorders in Type 2 diabetic mice and in palmitate-treated HepG2 cells. Eur. J. Pharmacol. 2016, 791, 254–263. [Google Scholar] [CrossRef]
- Willeit, P.; Skroblin, P.; Moschen, A.R.; Yin, X.; Kaudewitz, D.; Zampetaki, A.; Barwari, T.; Whitehead, M.; Ramírez, C.M.; Goedeke, L.; et al. Circulating MicroRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 2017, 66, 347–357. [Google Scholar] [CrossRef]
- Zhang, J.; Yao, T.; Wang, Y.; Yu, J.; Liu, Y.; Lin, Z. Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21. Cancer Biol. Ther. 2016, 17, 104–113. [Google Scholar] [CrossRef]
- You, L.; Wang, N.; Yin, D.; Wang, L.; Jin, F.; Zhu, Y.; Yuan, Q.; De, W. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J. Cell. Physiol. 2016, 231, 852–862. [Google Scholar] [CrossRef]
- Qiu, G.Z.; Tian, W.; Fu, H.T.; Li, C.P.; Liu, B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem. Biophys. Res. Commun. 2016, 471, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Zha, F.; Qu, X.; Tang, B.; Li, J.; Wang, Y.; Zheng, P.X.; Ji, T.; Zhu, C.; Bai, S. Long non-coding RNA MEG3 promotes fibrosis and inflammatory response in diabetic nephropathy via miR-181a/Egr-1/TLR4 axis. Aging 2019, 11, 3716–3730. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wu, Y.B.; Zhou, J.; Kang, D.M. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem. Biophys. Res. Commun. 2016, 469, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Shen, D.Y.; Han, C.K.; Tian, Y. LncRNA MEG3 aggravates palmitateinduced insulin resistance by regulating miR-185-5p/Egr2 axis in hepatic cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5456–5467. [Google Scholar] [PubMed]
- Sathishkumar, C.; Prabu, P.; Mohan, V.; Balasubramanyam, M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum. Genom. 2018, 12, 1–9. [Google Scholar] [CrossRef]
- Nilsson, E.; Matte, A.; Perfilyev, A.; De Mello, V.D.; Käkelä, P.; Pihlajamäki, J.; Ling, C. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J. Clin. Endocrinol. Metab. 2015, 100, E1491–E1501. [Google Scholar] [CrossRef]
- Zhang, N.; Geng, T.; Wang, Z.; Zhang, R.; Cao, T.; Camporez, J.P.; Cai, S.Y.; Liu, Y.; Dandolo, L.; Shulman, G.I.; et al. Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 2018, 3, 1–13. [Google Scholar] [CrossRef]
- Goyal, N.; Tiwary, S.; Kesharwani, D.; Datta, M. Long non-coding RNA H19 inhibition promotes hyperglycemia in mice by upregulating hepatic FoxO1 levels and promoting gluconeogenesis. J. Mol. Med. 2019, 97, 115–126. [Google Scholar] [CrossRef]
- Liu, J.; Tang, T.; Wang, G.D.; Liu, B. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARγ axis in non-alcoholic fatty liver disease. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Fawzy, M.; Abdelghany, A.; Toraih, E.; Mohamed, A. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosn. J. Basic Med. Sci. 2019, 8601. [Google Scholar] [CrossRef]
- Ahlman, B.; Charlton, M.; Fu, A.; Berg, C.; O’Brien, P.; Nair, K.S. Insulin’s effect on synthesis rates of liver proteins: A swine model comparing various precursors of protein synthesis. Diabetes 2001, 50, 947–954. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Q.; Lu, M.; Monks, B.R.; Birnbaum, M.J. Insulin is required to maintain albumin expression by inhibiting forkhead box O1 protein. J. Biol. Chem. 2016, 291, 2371–2378. [Google Scholar] [CrossRef] [PubMed]
- Biddinger, S.B.; Hernandez-Ono, A.; Rask-Madsen, C.; Haas, J.T.; Alemán, J.O.; Suzuki, R.; Scapa, E.F.; Agarwal, C.; Carey, M.C.; Stephanopoulos, G.; et al. Hepatic Insulin Resistance Is Sufficient to Produce Dyslipidemia and Susceptibility to Atherosclerosis. Cell Metab. 2008, 7, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, I.; Bashmakov, Y.; Ikemoto, S.; Horton, J.D.; Brown, M.S.; Goldstein, J.L. Insulin selectively increases SREBP-1C mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 1999, 96, 13656–13661. [Google Scholar] [CrossRef]
- Wu, X.; Williams, K.J. NOX4 pathway as a source of selective insulin resistance and responsiveness. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1236–1245. [Google Scholar] [CrossRef]
- Wu, X.; Chen, K.; Williams, K.J. The role of pathway-selective insulin resistance and responsiveness in diabetic dyslipoproteinemia. Curr. Opin. Lipidol. 2012, 23, 334–344. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, X.; Hu, S.; Yu, J. A Brief Review on the Mechanisms of miRNA Regulation. Genom. Proteom. Bioinforma. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Chang, H.Y. Long noncoding RNAs: Cellular address codes in development and disease. Cell 2013, 152, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Slavoff, S.A.; Mitchell, A.J.; Schwaid, A.G.; Cabili, M.N.; Ma, J.; Levin, J.Z.; Karger, A.D.; Budnik, B.A.; Rinn, J.L.; Saghatelian, A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013, 9, 59–64. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; Di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef]
- Liu, W.; Cao, H.; Yan, J.; Huang, R.; Ying, H. “Micro-managers” of hepatic lipid metabolism and NAFLD. Wiley Interdiscip. Rev. RNA 2015, 6, 581–593. [Google Scholar] [CrossRef]
- Goyal, N.; Kesharwani, D.; Datta, M. Lnc-ing non-coding RNAs with metabolism and diabetes: Roles of lncRNAs. Cell. Mol. Life Sci. 2018, 75, 1827–1837. [Google Scholar] [CrossRef]
- Huang, W.; Shi, Y.; Han, B.; Wang, Q.; Zhang, B.; Qi, C.; Liu, F. miR-802 inhibits the proliferation, invasion, and epithelial-mesenchymal transition of glioblastoma multiforme cells by directly targeting SIX4. Cell Biochem. Funct. 2020, 38, 66–76. [Google Scholar] [CrossRef]
- Wu, X.; Gong, Z.; Sun, L.; Ma, L.; Wang, Q. MicroRNA-802 plays a tumour suppressive role in tongue squamous cell carcinoma through directly targeting MAP2K4. Cell Prolif. 2017, 50, e12336. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Liu, X.; Pan, L.; Zhou, R.; Zhang, X. Long noncoding RNA MIR155HG facilitates pancreatic cancer progression through negative regulation of miR-802. J. Cell. Biochem. 2019, 120, 17926–17934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lv, R.; Guo, W.; Li, X. MicroRNA-802 inhibits cell proliferation and induces apoptosis in human cervical cancer by targeting serine/arginine-rich splicing factor 9. J. Cell. Biochem. 2019, 120, 10370–10379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Li, S.; Zhou, C.; Qin, Y.; Li, X. MiR-802 inhibits the aggressive behaviors of non-small cell lung cancer cells by directly targeting FGFR1. Int. J. Oncol. 2019, 54, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Zhao, Y.; Zhang, W.J.; Jiang, Y.J.; Fu, H.; Huang, F.; Li, D.J.; Shen, F.M. MicroRNA-802 accelerates hepatocellular carcinoma growth by targeting RUNX3. J. Cell. Physiol. 2020, jcp.29611. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, X.; Wang, M.; Lv, G.; Wang, G. High blood miR-802 is associated with poor prognosis in HCC patients by regulating DNA damage response 1 (REDD1)-mediated function of T cells. Oncol. Res. 2019, 27, 1025–1034. [Google Scholar] [CrossRef]
- Sun, D.; Chen, J.; Wu, W.; Tang, J.; Luo, L.; Zhang, K.; Jin, L.; Lin, S.; Gao, Y.; Yan, X.; et al. MiR-802 causes nephropathy by suppressing NF-κB-repressing factor in obese mice and human. J. Cell. Mol. Med. 2019, 23, 2863–2871. [Google Scholar] [CrossRef]
- Pessin, J.E.; Saltiel, A.R. Signaling pathways in insulin action: Molecular targets of insulin resistance. J. Clin. Invest. 2000, 106, 165–169. [Google Scholar] [CrossRef]
- Shieh, J.T.C.; Huang, Y.; Gilmore, J.; Srivastava, D. Elevated miR-499 levels blunt the cardiac stress response. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Z.; Fu, H.; Tie, Y.; Zhang, H.; Wu, Y.; Zheng, X. MicroRNA-1 and microRNA-499 downregulate the expression of the ets1 proto-oncogene in HepG2 cells. Oncol. Rep. 2012, 28, 701–706. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, R.; Zhang, J.; Li, W.; Gao, C.; Liu, J.; Wang, J. Identification of miR-423 and miR-499 polymorphisms on affecting the risk of hepatocellular carcinoma in a large-scale population. Genet. Test. Mol. Biomark. 2014, 18, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Ciccacci, C.; Latini, A.; Greco, C.; Politi, C.; D’Amato, C.; Lauro, D.; Novelli, G.; Borgiani, P.; Spallone, V. Association between a MIR499A polymorphism and diabetic neuropathy in type 2 diabetes. J. Diabetes Complicat. 2018, 32, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, T.; Chen, X.; Jiang, J.; Song, N.; Li, R.; Xin, Y.; Xuan, S. Inhibition of miR-499-5p expression improves nonalcoholic fatty liver disease. Ann. Hum. Genet. 2020, ahg.12374. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.L. Liver-specific microRNA-122: Biogenesis and function. RNA Biol. 2012, 9, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. MiR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef]
- Laudadio, I.; Manfroid, I.; Achouri, Y.; Schmidt, D.; Wilson, M.D.; Cordi, S.; Thorrez, L.; Knoops, L.; Jacquemin, P.; Schuit, F.; et al. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 2012, 142, 119–129. [Google Scholar] [CrossRef]
- Szabo, G.; Bala, S. MicroRNAs in liver disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 542–552. [Google Scholar] [CrossRef]
- Lowey, B.; Hertz, L.; Chiu, S.; Valdez, K.; Li, Q.; Liang, T.J. Hepatitis C virus infection induces hepatic expression of NF-κB-inducing kinase and lipogenesis by downregulating miR-122. MBio 2019, 10. [Google Scholar] [CrossRef]
- Li, C.; Deng, M.; Hu, J.; Li, X.; Chen, L.; Ju, Y.; Hao, J.; Meng, S. Chronic inflammation contributes to the development of hepatocellular carcinoma by decreasing miR-122 levels. The Oncotarget 2016, 7, 17021–17034. [Google Scholar] [CrossRef]
- Coulouarn, C.; Factor, V.M.; Andersen, J.B.; Durkin, M.E.; Thorgeirsson, S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. The Oncogene 2009, 28, 3526–3536. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, J.; Han, J.; Luo, D.; Sun, Z. MiR-122 inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Snail1 and Snail2 and suppressing WNT/β-cadherin signaling pathway. Exp. Cell Res. 2017, 360, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ke, S.; Zhong, L.; Wu, J.; Tseng, A.; Morpurgo, B.; Golovko, A.; Wang, G.; Cai, J.J.; Ma, X.; et al. Long noncoding RNA MALAT1 regulates generation of reactive oxygen species and the insulin responses in male mice. Biochem. Pharmacol. 2018, 152, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Eißmann, M.; Gutschner, T.; Hämmerle, M.; Günther, S.; Caudron-Herger, M.; Groß, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zörnig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell. Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Zhu, X.; Li, H.; Wu, Y.; Zhou, J.; Yang, G.; Wang, W. LncRNA MEG3 promotes hepatic insulin resistance by serving as a competing endogenous RNA of miR-214 to regulate ATF4 expression. Int. J. Mol. Med. 2019, 43, 345–357. [Google Scholar] [CrossRef]
- Goyal, N.; Sivadas, A.; Shamsudheen, K.V.; Jayarajan, R.; Verma, A.; Sivasubbu, S.; Scaria, V.; Datta, M. RNA sequencing of db/db mice liver identifies lncRNA H19 as a key regulator of gluconeogenesis and hepatic glucose output. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Church, R.J.; Otieno, M.; McDuffie, J.E.; Singh, B.; Sonee, M.; Hall, L.; Watkins, P.B.; Ellinger-Ziegelbauer, H.; Harrill, A.H. Beyond miR-122: Identification of MicroRNA Alterations in Blood During a Time Course of Hepatobiliary Injury and Biliary Hyperplasia in Rats. Toxicol. Sci. 2015, 150, 3–14. [Google Scholar] [CrossRef][Green Version]
- Wang, W.; Li, T.; Gao, L.; Li, Y.; Sun, Y.; Yao, H.-C. Plasma miR-208b and miR-499: Potential Biomarkers for Severity of Coronary Artery Disease. Dis. Markers 2019, 2019, 9842427. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.; Bao, X.; Wang, X.; Wu, J.; Li, X.; Hong, W. MiR-499-5p protects cardiomyocytes against ischaemic injury via anti-apoptosis by targeting PDCD4. The Oncotarget 2016, 7, 35607–35617. [Google Scholar] [CrossRef]
- Navickas, R.; Gal, D.; Laucevičius, A.; Taparauskaite, A.; Zdanyte, M.; Holvoet, P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: A systematic review. Cardiovasc. Res. 2016, 111, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Antonicelli, R.; Spazzafumo, L.; Santini, G.; Rippo, M.R.; Galeazzi, R.; Giovagnetti, S.; D’Alessandra, Y.; Marcheselli, F.; Capogrossi, M.C.; et al. Admission levels of circulating miR-499-5p and risk of death in elderly patients after acute non-ST elevation myocardial infarction. Int. J. Cardiol. 2014, 172, e276–e278. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Abu AlSel, B.T.; Al Ageeli, E.; Al-Qahtani, S.A.; Abdel-Daim, M.M.; Toraih, E.A. Long non-coding RNA MALAT1 and microRNA-499a expression profiles in diabetic ESRD patients undergoing dialysis: A preliminary cross-sectional analysis. Arch. Physiol. Biochem. 2020, 126, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Jampoka, K.; Muangpaisarn, P.; Khongnomnan, K.; Treeprasertsuk, S.; Tangkijvanich, P.; Payungporn, S. Serum miR-29a and miR-122 as Potential Biomarkers for Non-Alcoholic Fatty Liver Disease (NAFLD). Microrna 2018, 7, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Zhang, T.; Lou, G.; Xu, W.; Dong, F.; Chen, G.; Liu, Y. Plasma miR-17, miR-20a, miR-20b and miR-122 as potential biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus patients. Life Sci. 2018, 208, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Raitoharju, E.; Seppälä, I.; Lyytikäinen, L.P.; Viikari, J.; Ala-Korpela, M.; Soininen, P.; Kangas, A.J.; Waldenberger, M.; Klopp, N.; Illig, T.; et al. Blood hsa-MIR-122-5p and hsa-MIR-885-5p levels associate with fatty liver and related lipoprotein metabolism—The Young Finns Study. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Konishi, H.; Ichikawa, D.; Yamamoto, Y.; Arita, T.; Shoda, K.; Hiramoto, H.; Hamada, J.; Itoh, H.; Fujita, Y.; Komatsu, S.; et al. Plasma level of metastasis-associated lung adenocarcinoma transcript 1 is associated with liver damage and predicts development of hepatocellular carcinoma. Cancer Sci. 2016, 107, 149–154. [Google Scholar] [CrossRef]
- Toraih, E.A.; Ellawindy, A.; Fala, S.Y.; Al Ageeli, E.; Gouda, N.S.; Fawzy, M.S.; Hosny, S. Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed. Pharmacother. 2018, 102, 653–669. [Google Scholar] [CrossRef]
- Chen, M.J.; Wang, X.G.; Sun, Z.X.; Liu, X.C. Diagnostic value of LncRNA-MEG3 as a serum biomarker in patients with hepatitis B complicated with liver fibrosis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4360–4367. [Google Scholar] [PubMed]
- Dong, H.; Zhang, Y.; Xu, Y.; Ma, R.; Liu, L.; Luo, C.; Jiang, W. Downregulation of long non-coding RNA MEG3 promotes proliferation, migration, and invasion of human hepatocellular carcinoma cells by upregulating TGF-β1. Acta Biochim. Biophys. Sin. 2019, 51, 645–652. [Google Scholar] [CrossRef]
- Fawzy, F. Long Non-Coding RNA H19 as Potential Biomarker for HCV Genotype 4 Induced Hepatocellular Carcinoma Patients. Al Azhar J. Pharm. Sci. 2019, 60, 76–94. [Google Scholar] [CrossRef]
- Tello-Flores, V.A.; Valladares-Salgado, A.; Ramírez-Vargas, M.A.; Cruz, M.; del-Moral-Hernández, O.; Cahua-Pablo, J.Á.; Ramírez, M.; Hernández-Sotelo, D.; Armenta-Solis, A.; Flores-Alfaro, E. Altered levels of MALAT1 and H19 derived from serum or serum exosomes associated with type-2 diabetes. Non Coding RNA Res. 2020, 5, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Ghaedi, H.; Zare, A.; Omrani, M.D.; Doustimotlagh, A.H.; Meshkani, R.; Alipoor, S.; Alipoor, B. Genetic variants in long noncoding RNA H19 and MEG3 confer risk of type 2 diabetes in an Iranian population. Gene 2018, 675, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Franko, A.; Neschen, S.; Rozman, J.; Rathkolb, B.; Aichler, M.; Feuchtinger, A.; Brachthäuser, L.; Neff, F.; Kovarova, M.; Wolf, E.; et al. Bezafibrate ameliorates diabetes via reduced steatosis and improved hepatic insulin sensitivity in diabetic TallyHo mice. Mol. Metab. 2017, 6, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Alkhalidy, H.; Moore, W.; Wang, A.; Luo, J.; McMillan, R.P.; Wang, Y.; Zhen, W.; Hulver, M.W.; Liu, D. Kaempferol ameliorates hyperglycemia through suppressing hepatic gluconeogenesis and enhancing hepatic insulin sensitivity in diet-induced obese mice. J. Nutr. Biochem. 2018, 58, 90–101. [Google Scholar] [CrossRef]
- Sharma, R.; Matsuzaka, T.; Kaushik, M.K.; Sugasawa, T.; Ohno, H.; Wang, Y.; Motomura, K.; Shimura, T.; Okajima, Y.; Mizunoe, Y.; et al. Octacosanol and policosanol prevent high-fat diet-induced obesity and metabolic disorders by activating brown adipose tissue and improving liver metabolism. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Leidinger, P.; Backes, C.; Deutscher, S.; Schmitt, K.; Mueller, S.C.; Frese, K.; Haas, J.; Ruprecht, K.; Paul, F.; Stähler, C.; et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 2013, 14, 1–16. [Google Scholar] [CrossRef]
- Zhang, K.; Luo, Z.; Zhang, Y.; Zhang, L.; Wu, L.; Liu, L.; Yang, J.; Song, X.; Liu, J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016, 17, 187–194. [Google Scholar] [CrossRef]
- Heneghan, H.M.; Miller, N.; Kelly, R.; Newell, J.; Kerin, M.J. Systemic miRNA-195 Differentiates Breast Cancer from Other Malignancies and Is a Potential Biomarker for Detecting Noninvasive and Early Stage Disease. The Oncologist 2010, 15, 673–682. [Google Scholar] [CrossRef]
- Gu, Y.; Chen, T.; Li, G.; Yu, X.; Lu, Y.; Wang, H.; Teng, L. LncRNAs: Emerging biomarkers in gastric cancer. Futur. Oncol. 2015, 11, 2427–2441. [Google Scholar] [CrossRef]
- DIng, H.; Meng, J.; Zhang, W.; Li, Z.; Li, W.; Zhang, M.; Fan, Y.; Wang, Q.; Zhang, Y.; Jiang, L.; et al. Medical examination powers miR-194-5p as a biomarker for postmenopausal osteoporosis. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gui, T.; Shen, K. MiRNA-101: A potential target for tumor therapy. Cancer Epidemiol. 2012, 36, 537–540. [Google Scholar] [CrossRef] [PubMed]
| ncRNA | Status in Liver | Status in Circulation | Reference |
|---|---|---|---|
| miR-802 | ↑ | ↑ | [22,24,25] |
| miR-499-5p | ↓ | ↓ | [26,27] |
| miR-122-5p | ↑ | ↑ | [28,29,30,31,32] |
| lnc MEG3 | ↑ | ↑ | [33,34,35,36] |
| lnc MALAT1 | ↑ | ↑ | [23,37,38,39] |
| lnc H19 | ↑/↓ * | ↑ | [40,41,42,43,44] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pielok, A.; Marycz, K. Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance. Int. J. Mol. Sci. 2020, 21, 4182. https://doi.org/10.3390/ijms21114182
Pielok A, Marycz K. Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance. International Journal of Molecular Sciences. 2020; 21(11):4182. https://doi.org/10.3390/ijms21114182
Chicago/Turabian StylePielok, Ariadna, and Krzysztof Marycz. 2020. "Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance" International Journal of Molecular Sciences 21, no. 11: 4182. https://doi.org/10.3390/ijms21114182
APA StylePielok, A., & Marycz, K. (2020). Non-Coding RNAs as Potential Novel Biomarkers for Early Diagnosis of Hepatic Insulin Resistance. International Journal of Molecular Sciences, 21(11), 4182. https://doi.org/10.3390/ijms21114182





