Insight into the Regulatory Relationships between the Insulin-Like Androgenic Gland Hormone Gene and the Insulin-Like Androgenic Gland Hormone-binding Protein Gene in Giant Freshwater Prawns (Macrobrachium rosenbergii)
Abstract
:1. Introduction
2. Results
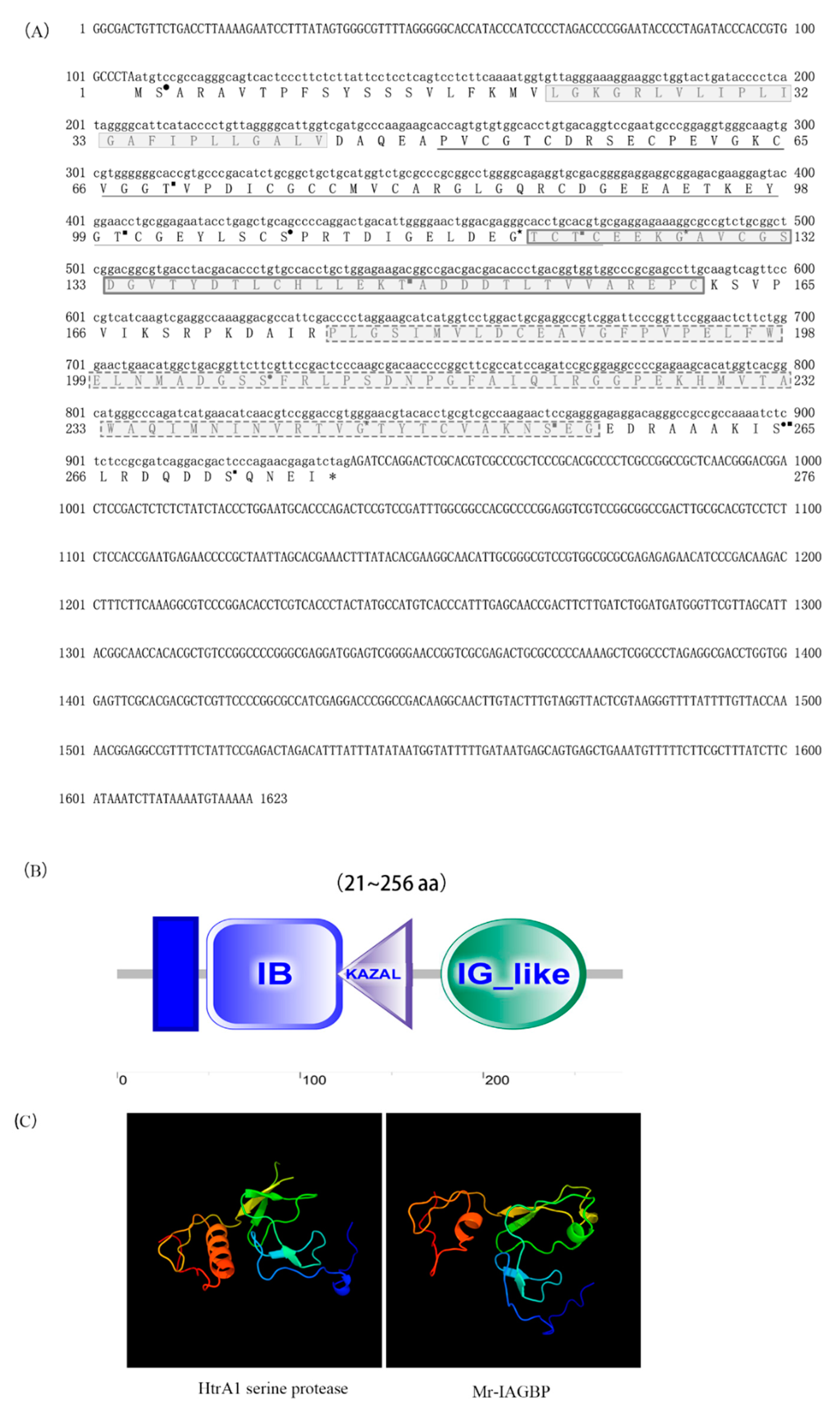
2.1. Isolation of Full-Length Mr-IAGBP cDNA
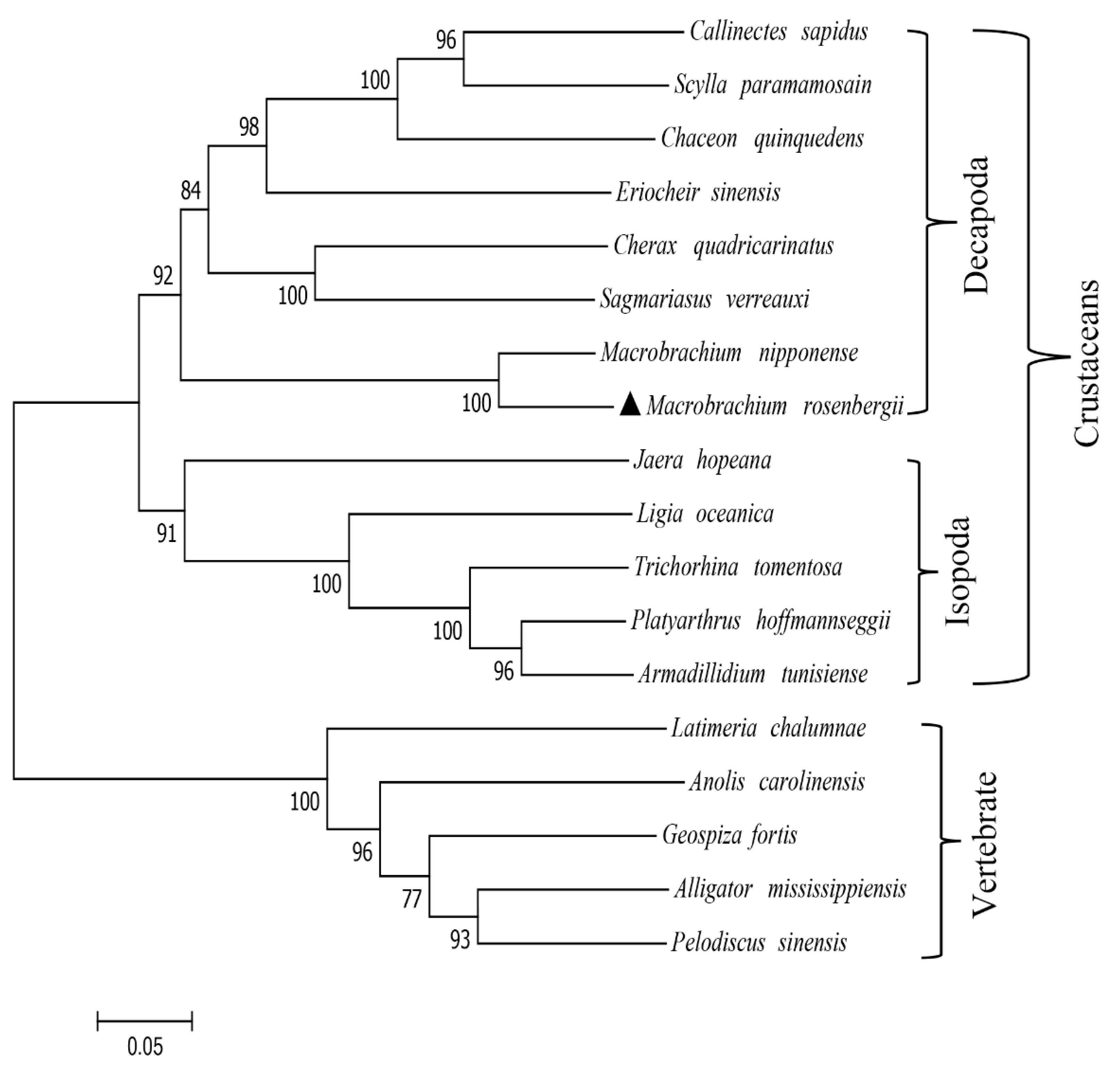
2.2. Multiple Alignment and Phylogenetic Analysis
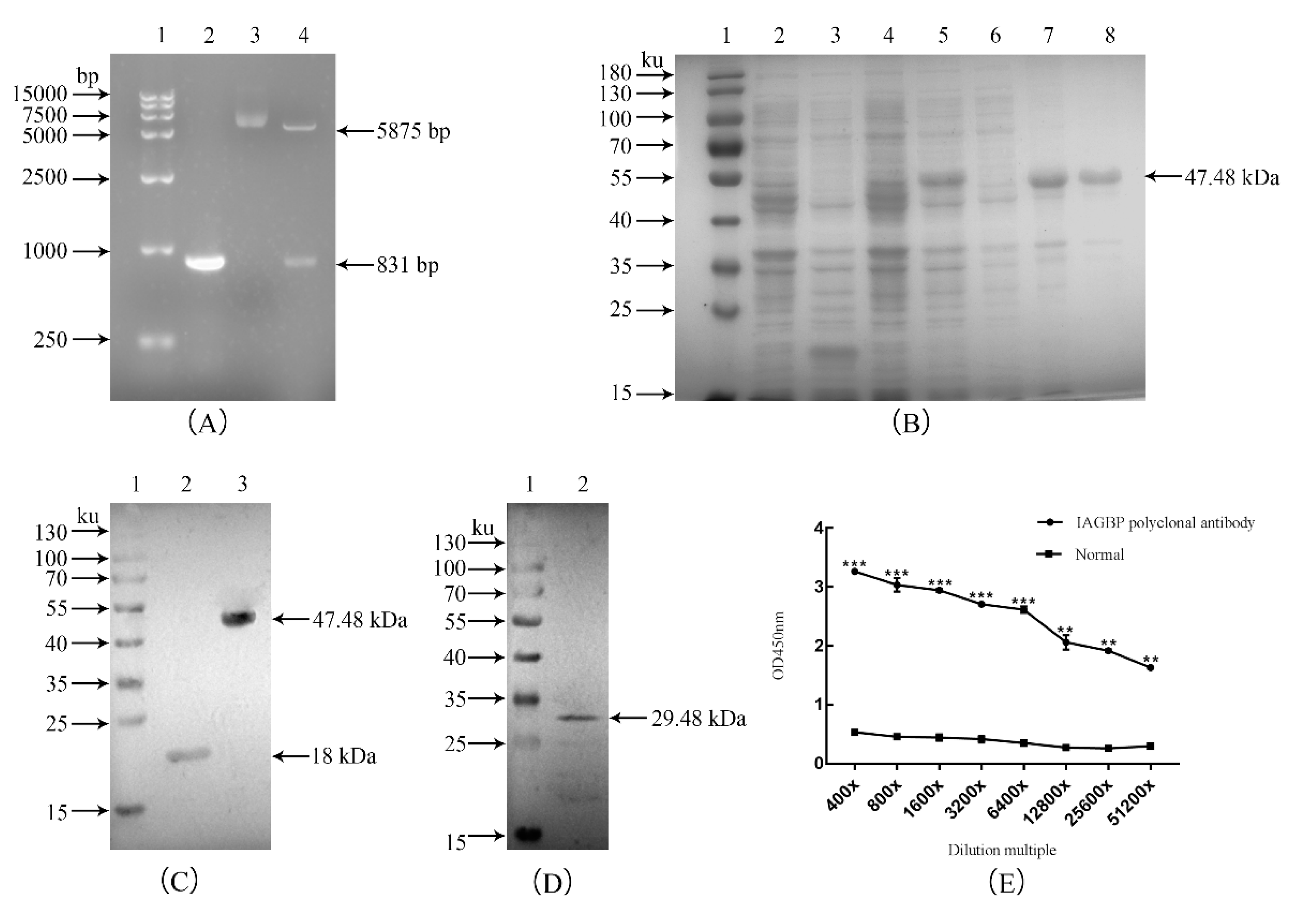
2.3. Recombinant Mr-IAGBP Protein Expression, Purification, and Polyclonal Antibodies Analysis
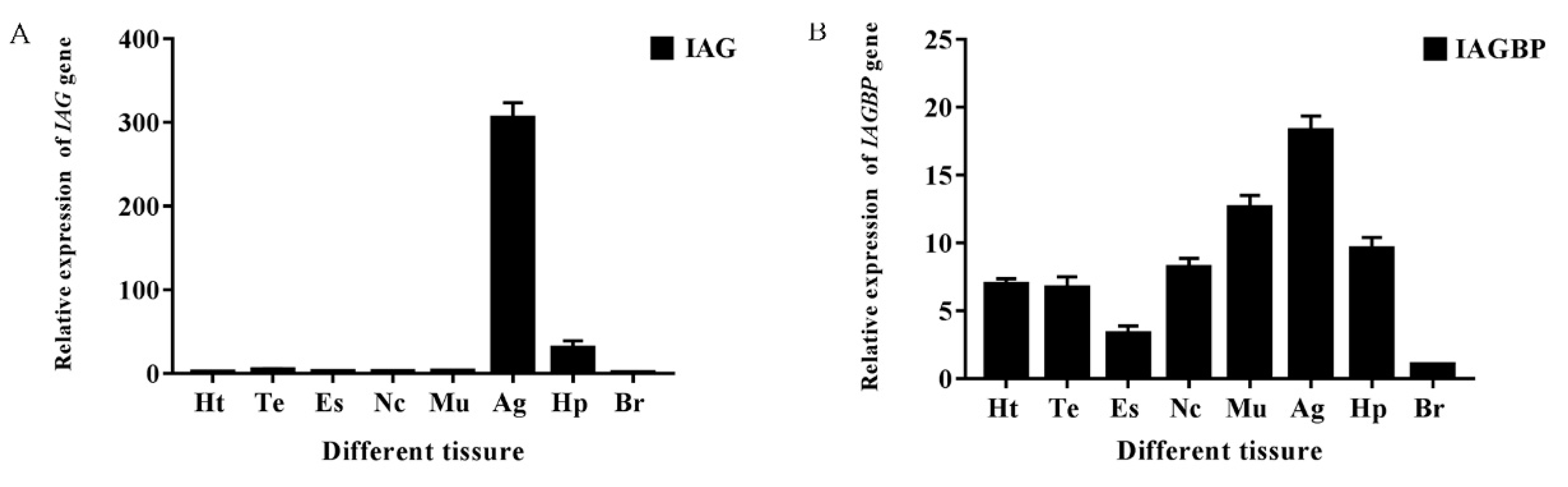
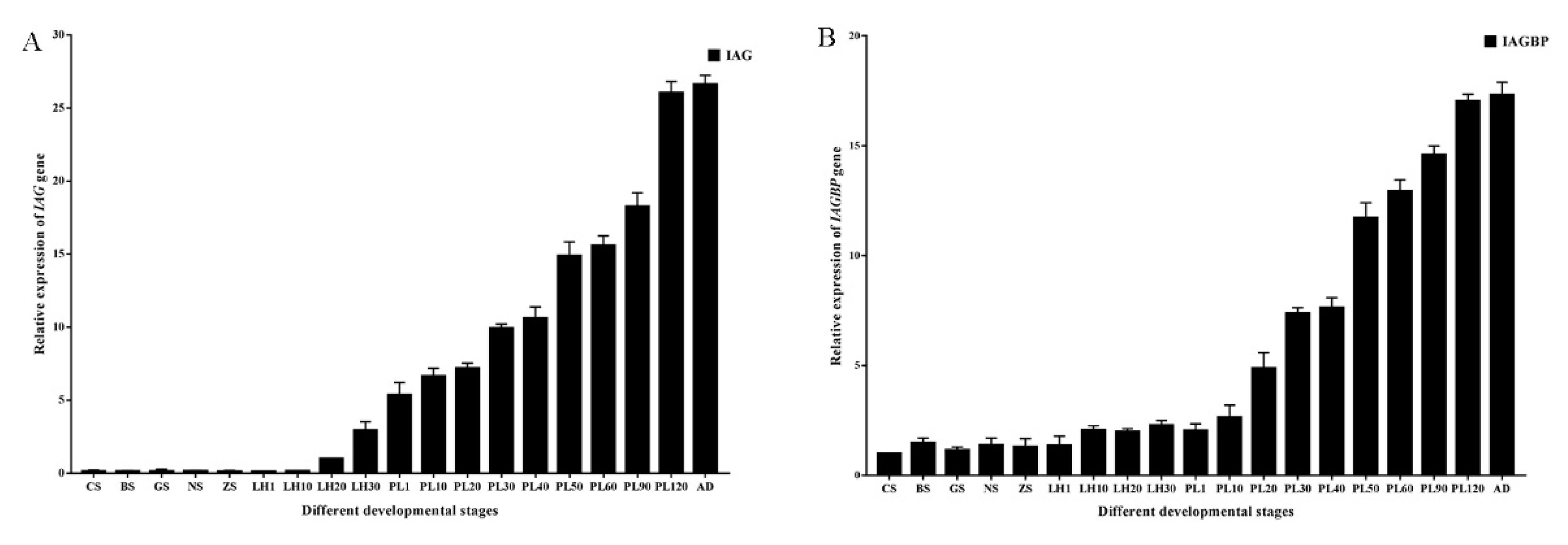
2.4. Tissue and Spatial Distributions of Mr-IAGBP and Mr-IAG Transcripts
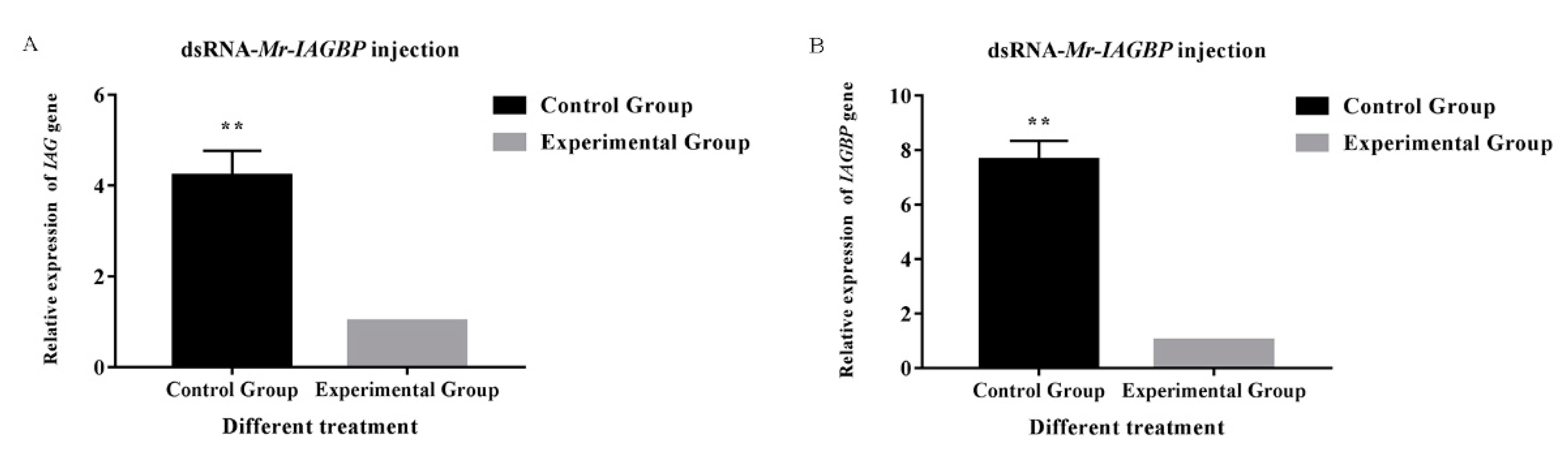
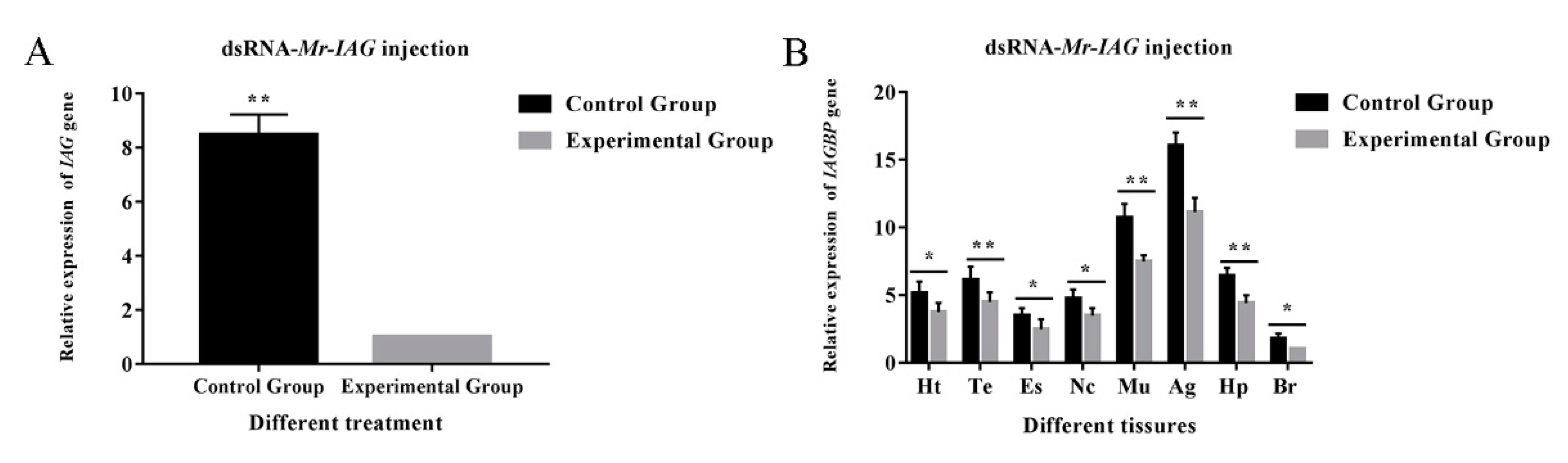
2.5. Transcription in Mr-IAG-dsRNA- and Mr-IAGBP-dsRNA-injection Groups
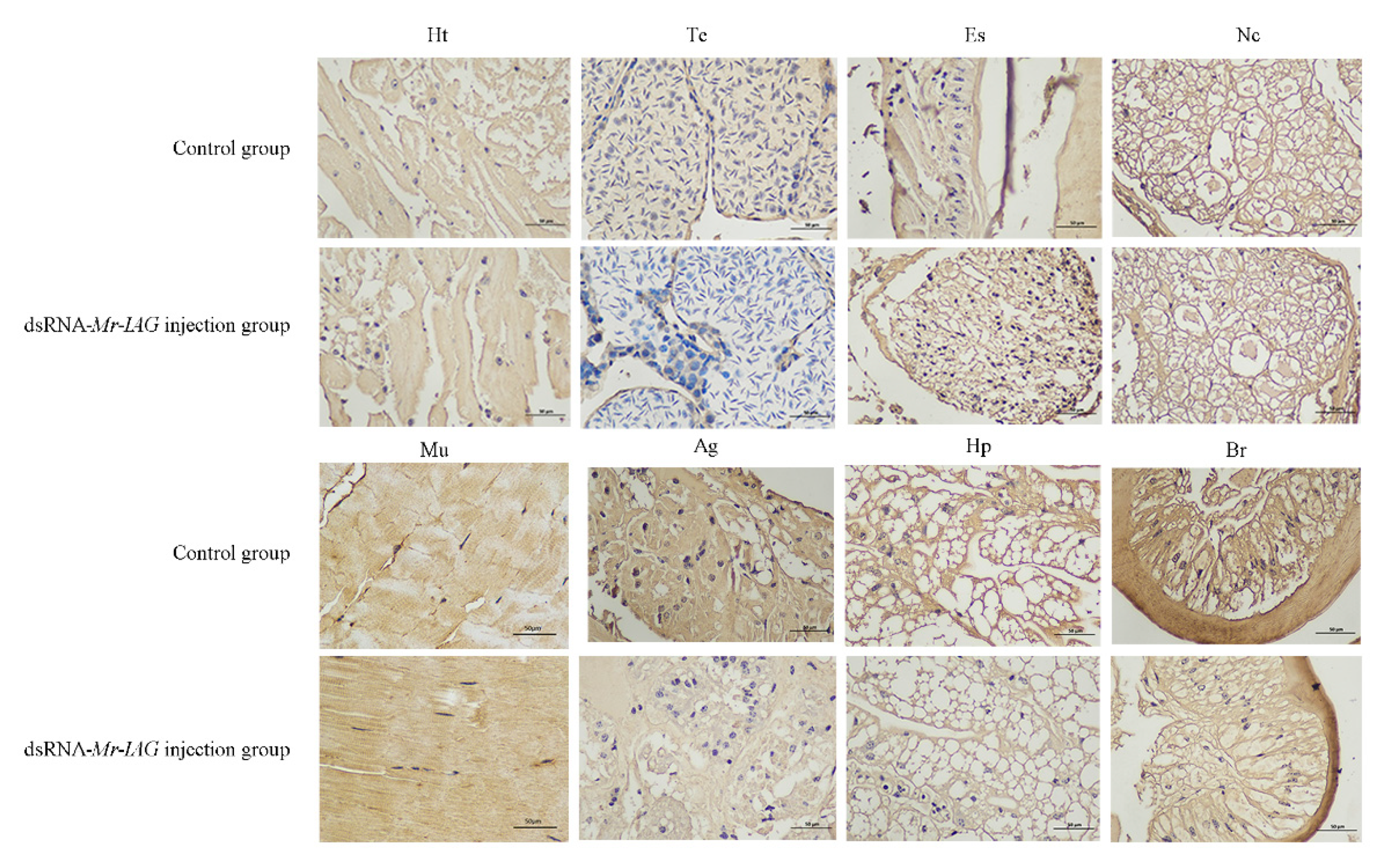
2.6. Western Blotting and Immunohistochemistry Analyses
3. Discussion
4. Materials and Methods
4.1. Tissue Sampling, RNA Isolation, and Reverse Transcription
4.2. Gene Cloning of Mr-IAGBP and Bioinformatics Analyses
4.3. Construction of Recombinant Mr-IAGBP Plasmid, Expressing and Purification
4.4. Rabbit Polyclonal Antibodies Against Recombinant Mr-IAGBP and Western-blot
4.5. qRT-PCR Analysis of Mr-IAGBP and Mr-IAG
4.6. Double-Stranded RNA Preparation
4.7. In Vivo Mr-IAG and Mr-IAGBP Silencing
4.8. Immunohistochemistry Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AG | Androgenic gland |
| IGFBP | Insulin-like growth factor-binding protein |
| ORF | Open reading frame |
| PBS | Phosphate buffer saline |
| TBST | Tris buffered saline tween |
| HRP | Horseradish Peroxidase |
| BSA | Bovine serum albumin |
| EDTA | Ethylene diamine tetraacetic acid |
| DEPC | Diethyl pyrocarbonate |
References
- New, M.B. Freshwater prawn farming: Global status, recent research and a glance at the future. Aquac. Res. 2005, 36, 210–230. [Google Scholar] [CrossRef]
- Hartnoll, R.G. Growth in Crustacea – twenty years on. Hydrobiologia 2001, 449, 111–122. [Google Scholar] [CrossRef]
- Sagi, A.; Ra’anan, Z.; Cohen, D.; Wax, Y. Production of Macrobrachium rosenbergii in monosex populations: Yield characteristics under intensive monoculture conditions in cages. Aquaculture 1986, 51, 265–275. [Google Scholar] [CrossRef]
- Cronin, L.E. Anatomy and histology of the male reproductive system of Callinectes sapidus Rathbun. J. Morphol. 1947, 81, 209–239. [Google Scholar] [CrossRef]
- Aflalo, E.; Hoang, T.T.T.; Nguyen, V.H.; Lam, Q.; Nguyen, D.M.; Trinh, Q.S.; Raviv, S.; Sagi, A. A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2006, 256, 468–478. [Google Scholar] [CrossRef]
- Barki, A.; Karplus, I.; Khalaila, I.; Manor, R.; Sagi, A. Male-like behavioral patterns and physiological alterations induced by androgenic gland implantation in female crayfish. J. Exp. Biol. 2003, 206, 1791–1797. [Google Scholar] [CrossRef] [Green Version]
- Nagamine, C.; Knight, A.W. Induction of Female Breeding Characteristics by Ovarian Tissue Implants in Androgenic Gland Ablated Male Freshwater Prawns Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae). Int. J. Invertebr. Reprod. Dev. 1987, 11, 225–234. [Google Scholar] [CrossRef]
- Nagamine, C.; Knight, A.W.; Maggenti, A.; Paxman, G. Effects of androgenic gland ablation on male primary and secondary sexual characteristics in the Malaysian prawn, Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae), with first evidence of induced feminization in a nonhermaphroditic decapod. Gen. Comp. Endocrinol. 1980, 41, 423–441. [Google Scholar] [CrossRef]
- Nagamine, C.; Knight, A.W.; Maggenti, A.; Paxman, G. Masculinization of female Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae) by androgenic gland implantation. Gen. Comp. Endocrinol. 1980, 41, 442–457. [Google Scholar] [CrossRef]
- Sagi, A.; Cohen, D.; Milner, Y. Effect of androgenic gland ablation on morphotypic differentiation and sexual characteristics of male freshwater prawns, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 1990, 77, 15–22. [Google Scholar] [CrossRef]
- Abdu, U.; Davis, C.; Khalaila, I.; Sagi, A. The vitellogenin cDNA of Cherax quadricarinatus encodes a lipoprotein with calcium binding ability, and its expression is induced following the removal of the androgenic gland in a sexually plastic system. Gen. Comp. Endocrinol. 2002, 127, 263–272. [Google Scholar] [CrossRef]
- Barki, A.; Karplus, I.; Manor, R.; Sagi, A. Intersexuality and behavior in crayfish: The de-masculinization effects of androgenic gland ablation. Horm. Behav. 2006, 50, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, F.; Sun, Z.; Xiang, J. Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis. Gen. Comp. Endocrinol. 2012, 177, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Manor, R.; Aflalo, E.; Segall, C.; Weil, S.; Azulay, D.; Ventura, T.; Sagi, A. Androgenic gland implantation promotes growth and inhibits vitellogenesis in Cherax quadricarinatus females held in individual compartments. Invertebr. Reprod. Dev. 2004, 45, 151–159. [Google Scholar] [CrossRef]
- Manor, R.; Weil, S.; Oren, S.; Glazer, L.; Aflalo, E.D.; Ventura, T.; Chalifa-Caspi, V.; Lapidot, M.; Sagi, A. Insulin and gender: An insulin-like gene expressed exclusively in the androgenic gland of the male crayfish. Gen. Comp. Endocrinol. 2007, 150, 326–336. [Google Scholar] [CrossRef]
- Mareddy, V.; Rosen, O.; Thaggard, H.B.; Manor, R.; Kuballa, A.; Aflalo, E.; Sagi, A.; Paterson, B.; Elizur, A. Isolation and characterization of the complete cDNA sequence encoding a putative insulin-like peptide from the androgenic gland of Penaeus monodon. Aquaculture 2011, 318, 364–370. [Google Scholar] [CrossRef]
- Ohira, T.; Hasegawa, Y.; Tominaga, S.; Okuno, A.; Nagasawa, H. Molecular cloning and expression analysis of cDNAs encoding androgenic gland hormone precursors from two porcellionidae species, Porcellio scaber and P. dilatatus. Zool. Sci. 2003, 20, 75–81. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2009, 150, 1278–1286. [Google Scholar] [CrossRef] [Green Version]
- Ma, K.Y.; Lin, J.Y.; Guo, S.Z.; Chen, Y.; Li, J.L.; Qiu, G.F. Molecular characterization and expression analysis of an insulin-like gene from the androgenic gland of the oriental river prawn, Macrobrachium nipponense. Gen. Comp. Endocrinol. 2013, 185, 90–96. [Google Scholar] [CrossRef]
- Ventura, T.; Fitzgibbon, Q.; Battaglene, S.; Sagi, A.; Elizur, A. Identification and characterization of androgenic gland specific insulin-like peptide-encoding transcripts in two spiny lobster species: Sagmariasus verreauxi and Jasus edwardsii. Gen. Comp. Endocrinol. 2015, 214, 126–133. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, K.; Wang, S.; Peng, H.; Shan, Z.; Wang, K. Molecular identification of a new androgenic gland-specific insulin-like gene from the mud crab, Scylla paramamosain. Aquaculture 2014, 433, 325334. [Google Scholar] [CrossRef]
- Li, F.; Bai, H.; Xiong, Y.; Fu, H.; Jiang, S.; Jiang, F.; Jin, S.; Sun, S.; Qiao, H.; Zhang, W. Molecular characterization of insulin-like androgenic gland hormone-binding protein gene from the oriental river prawn Macrobrachium nipponense and investigation of its transcriptional relationship with the insulin-like androgenic gland hormone gene. Gen. Comp. Endocrinol. 2015, 216, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Rosen, O.; Weil, S.; Manor, R.; Roth, Z.; Khalaila, I.; Sagi, A. A crayfish insulin-like-binding protein: Another piece in the androgenic gland insulin-like hormone puzzle is revealed. J. Biol. Chem. 2013, 288, 22289–22298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandler, J.C.; Aizen, J.; Elizur, A.; Hollander-Cohen, L.; Battaglene, S.C.; Ventura, T. Discovery of a novel insulin-like peptide and insulin binding proteins in the Eastern rock lobster Sagmariasus verreauxi. Gen. Comp. Endocrinol. 2015, 215, 76–87. [Google Scholar] [CrossRef]
- Kruangkum, T.; Saetan, J.; Chotwiwatthanakun, C.; Vanichviriyakit, R.; Thongrod, S.; Thintharua, P.; Tulyananda, T.; Sobhon, P. Co-culture of males with late premolt to early postmolt female giant freshwater prawns, Macrobrachium rosenbergii resulted in greater abundances of insulin-like androgenic gland hormone and gonad maturation in male prawns as a result of olfactory receptors. Anim. Reprod. Sci. 2019, 210, 106198. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Khalaila, I.; Rosen, O.; Sagi, A. Expression of an Androgenic Gland-Specific Insulin-Like Peptide during the Course of Prawn Sexual and Morphotypic Differentiation. Isrn Endocrinol. 2011, 2011, 476283. [Google Scholar] [CrossRef] [Green Version]
- Shpak, N.; Manor, R.; Abilevich, L.K.; Mantal, O.; Shavit, K.; Aflalo, E.D.; Toiber, D.; Sagi, A. Short versus long double-stranded RNA activation of a post-transcriptional gene knockdown pathway. Rna Biol. 2017, 14, 1766–1775. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 90. [Google Scholar] [CrossRef]
- Priyadarshi, H.; Das, R.; Pavan-Kumar, A.; Gireesh-Babu, P. Silencing and augmentation of IAG hormone transcripts in adult Macrobrachium rosenbergii males affects morphotype transformation. J. Exp. Biol. 2017, 220, 4101–4108. [Google Scholar] [CrossRef] [Green Version]
- Phoungpetchara, I.; Tinikul, Y.; Poljaroen, J.; Chotwiwatthanakun, C.; Vanichviriyakit, R.; Sroyraya, M.; Hanna, P.J.; Sobhon, P. Cells producing insulin-like androgenic gland hormone of the giant freshwater prawn, Macrobrachium rosenbergii, proliferate following bilateral eyestalk-ablation. Tissue Cell 2011, 43, 165–177. [Google Scholar] [CrossRef]
- Siangcham, T.; Tinikul, Y.; Poljaroen, J.; Sroyraya, M.; Changklungmoa, N.; Phoungpetchara, I.; Kankuan, W.; Sumpownon, C.; Wanichanon, C.; Hanna, P.J.; et al. The effects of serotonin, dopamine, gonadotropin-releasing hormones, and corazonin, on the androgenic gland of the giant freshwater prawn, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 2013, 193, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Daza, D.O.; Sundstrom, G.; Bergqvist, C.A.; Duan, C.; Larhammar, D. Evolution of the insulin-like growth factor binding protein (IGFBP) family. Endocrinology 2011, 152, 2278–2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwa, V.; oh, Y.; Rosenfeld, R. The Insulin-Like Growth Factor-Binding Protein (IGFBP) Superfamily 1. Endocr. Rev. 2000, 20, 761–787. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in crayfish: Ovarian upregulation and testicular degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharabi, O.; Manor, R.; Weil, S.; Aflalo, E.D.; Lezer, Y.; Levy, T.; Aizen, J.; Ventura, T.; Mather, P.B.; Khalaila, I.; et al. Identification and Characterization of an Insulin-Like Receptor Involved in Crustacean Reproduction. Endocrinology 2016, 157, 928–941. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Li, S.; Lv, X.; Xiang, J.; Sagi, A.; Manor, R.; Li, F. A Putative Insulin-like Androgenic Gland Hormone Receptor Gene Specifically Expressed in Male Chinese Shrimp. Endocrinology 2018, 159, 2173–2185. [Google Scholar] [CrossRef] [Green Version]
- Habashy, M.M.; Sharshar, K.M.; Hassan, M.M.S. Morphological and histological studies on the embryonic development of the freshwater prawn, Macrobrachium rosenbergii (Crustacea, Decapoda). J. Basic Appl. Zool. 2012, 65, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Tinikul, Y.; Poljaroen, J.; Tinikul, R.; Sobhon, P. Changes in the levels, expression, and possible roles of serotonin and dopamine during embryonic development in the giant freshwater prawn, Macrobrachium rosenbergii. Gen. Comp. Endocrinol. 2016, 225, 71–80. [Google Scholar] [CrossRef]
- Tinikul, Y.; Soonthornsumrith, B.; Phoungpetchara, I.; Meeratana, P.; Poljaroen, J.; Duangsuwan, P.; Soonklang, N.; Mercier, J.; Sobhon, P. Effects of serotonin, dopamine, octopamine, and spiperone on ovarian maturation and embryonic development in the giant freshwater prawn, Macrobrachium rosenbergii (De Man, 1879). Crustaceana 2009, 82, 1007–1022. [Google Scholar]
- Amterat Abu Abayed, F.; Manor, R.; Aflalo, E.D.; Sagi, A. Screening for Dmrt genes from embryo to mature Macrobrachium rosenbergii prawns. Gen. Comp. Endocrinol. 2019, 282, 113205. [Google Scholar] [CrossRef]
- Borisov, R.R.; Kriakhova, N.V. Influence of lecithotrofic feeding on growth and development of larvae of freshwater shrimp Macrobrachium rosenbergii. Ontogenez 2011, 42, 178–182. [Google Scholar] [PubMed]
- Lu, Z.; Qin, Z.; Babu, S.; Ye, C.; Su, G.; Li, J.; Yang, G.; Shen, H.; Pan, G.; Lin, L. Expression and functional characterization of glutamine synthetase from giant freshwater prawn (Macrobrachium rosenbergii ) under osmotic stress. Aquac. Res. 2019, 50, 2635–2645. [Google Scholar] [CrossRef]
- Lu, Z.; Yang, G.; Qin, Z.; Shen, H.; Zhang, M.; Shi, F.; Li, J.; Sarath Babu, V.; Lin, L. Glutamate related osmoregulation of guanine nucleotide-binding protein G (I) α2 from giant freshwater prawn (Macrobrachium rosenbergii) during molting and salinity stress. Aquaculture 2020, 521, 735000. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Wu, C.; Zhao, X.; Babu, V.S.; Yuan, G.; Wang, W.; Su, J.; Liu, X.; Lin, L. Distribution of mannose receptor in blunt snout bream (Megalobrama amblycephala) during the embryonic development and its immune response to the challenge of Aeromonas hydrophila. Fish. Shellfish Immunol. 2018, 78, 52–59. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Lu, Z.; Qin, Z.; Zhao, L.; Pan, G.; Shen, H.; Zhang, M.; Liang, R.; Lin, L.; Zhang, K. Insight into the Regulatory Relationships between the Insulin-Like Androgenic Gland Hormone Gene and the Insulin-Like Androgenic Gland Hormone-binding Protein Gene in Giant Freshwater Prawns (Macrobrachium rosenbergii). Int. J. Mol. Sci. 2020, 21, 4207. https://doi.org/10.3390/ijms21124207
Yang G, Lu Z, Qin Z, Zhao L, Pan G, Shen H, Zhang M, Liang R, Lin L, Zhang K. Insight into the Regulatory Relationships between the Insulin-Like Androgenic Gland Hormone Gene and the Insulin-Like Androgenic Gland Hormone-binding Protein Gene in Giant Freshwater Prawns (Macrobrachium rosenbergii). International Journal of Molecular Sciences. 2020; 21(12):4207. https://doi.org/10.3390/ijms21124207
Chicago/Turabian StyleYang, Guang, Zhijie Lu, Zhendong Qin, Lijuan Zhao, Gan Pan, Haiyang Shen, Menglan Zhang, Rishen Liang, Li Lin, and Kai Zhang. 2020. "Insight into the Regulatory Relationships between the Insulin-Like Androgenic Gland Hormone Gene and the Insulin-Like Androgenic Gland Hormone-binding Protein Gene in Giant Freshwater Prawns (Macrobrachium rosenbergii)" International Journal of Molecular Sciences 21, no. 12: 4207. https://doi.org/10.3390/ijms21124207
APA StyleYang, G., Lu, Z., Qin, Z., Zhao, L., Pan, G., Shen, H., Zhang, M., Liang, R., Lin, L., & Zhang, K. (2020). Insight into the Regulatory Relationships between the Insulin-Like Androgenic Gland Hormone Gene and the Insulin-Like Androgenic Gland Hormone-binding Protein Gene in Giant Freshwater Prawns (Macrobrachium rosenbergii). International Journal of Molecular Sciences, 21(12), 4207. https://doi.org/10.3390/ijms21124207




