GLI1 and AXIN2 Are Distinctive Markers of Human Calvarial Mesenchymal Stromal Cells in Nonsyndromic Craniosynostosis
Abstract
:1. Introduction
2. Results
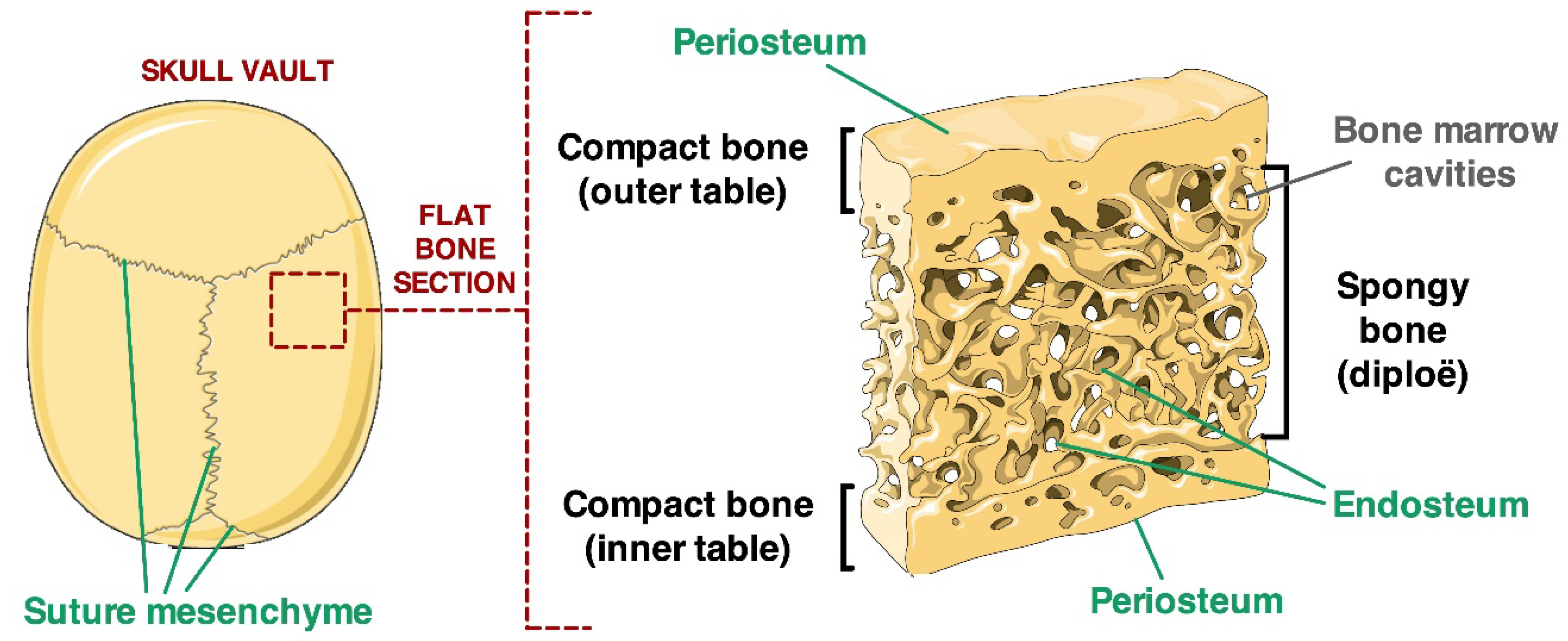
2.1. Localisation of Calvarial Stem Cell Markers in Suture Tissue Samples
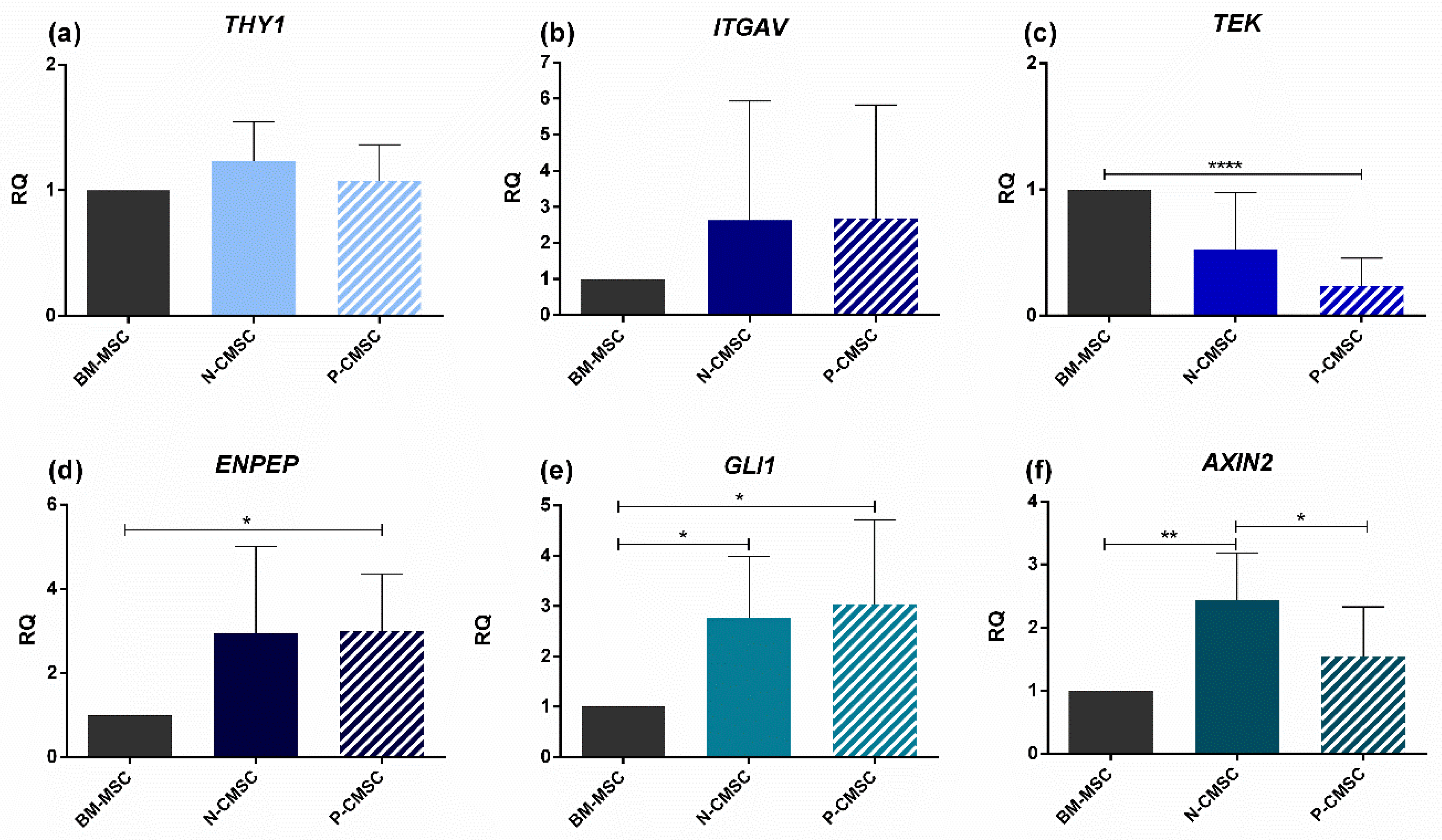
2.2. Expression of Calvarial Stem Cell Markers in Calvarial Mesenchymal Stromal Cells (CMSC)
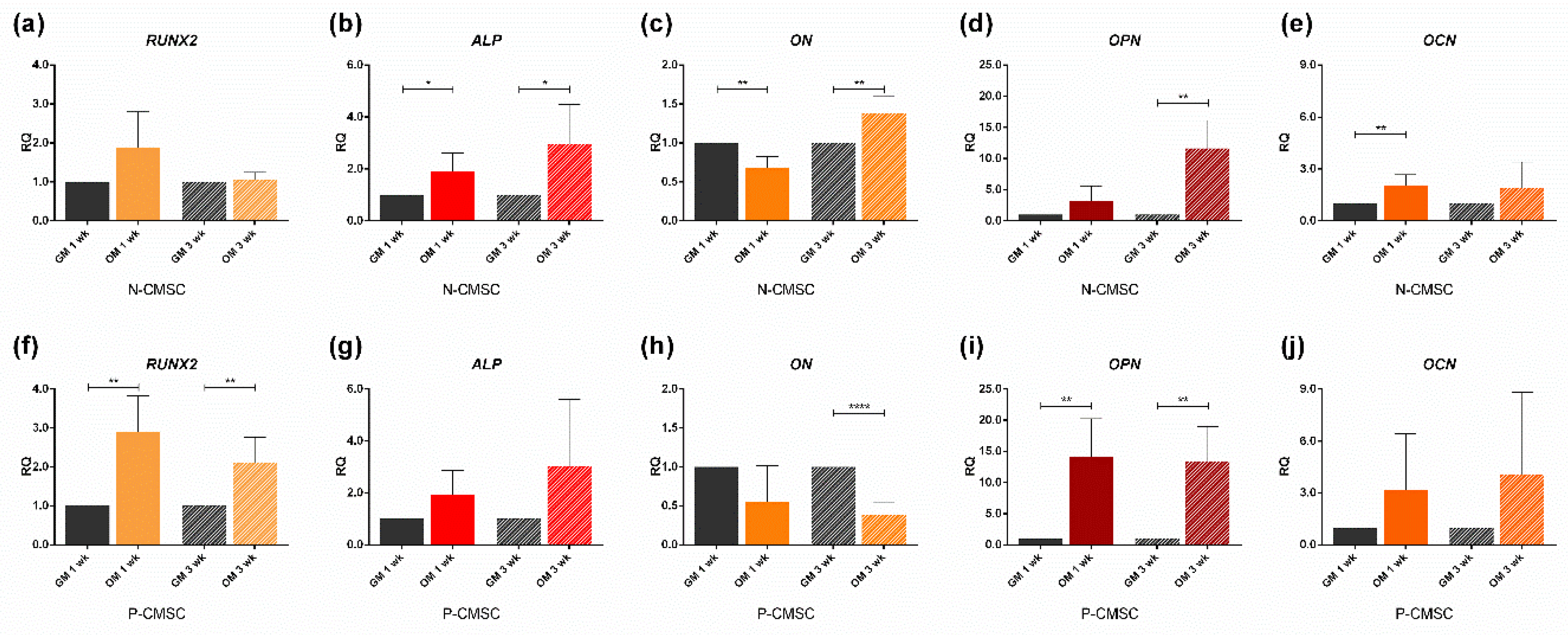
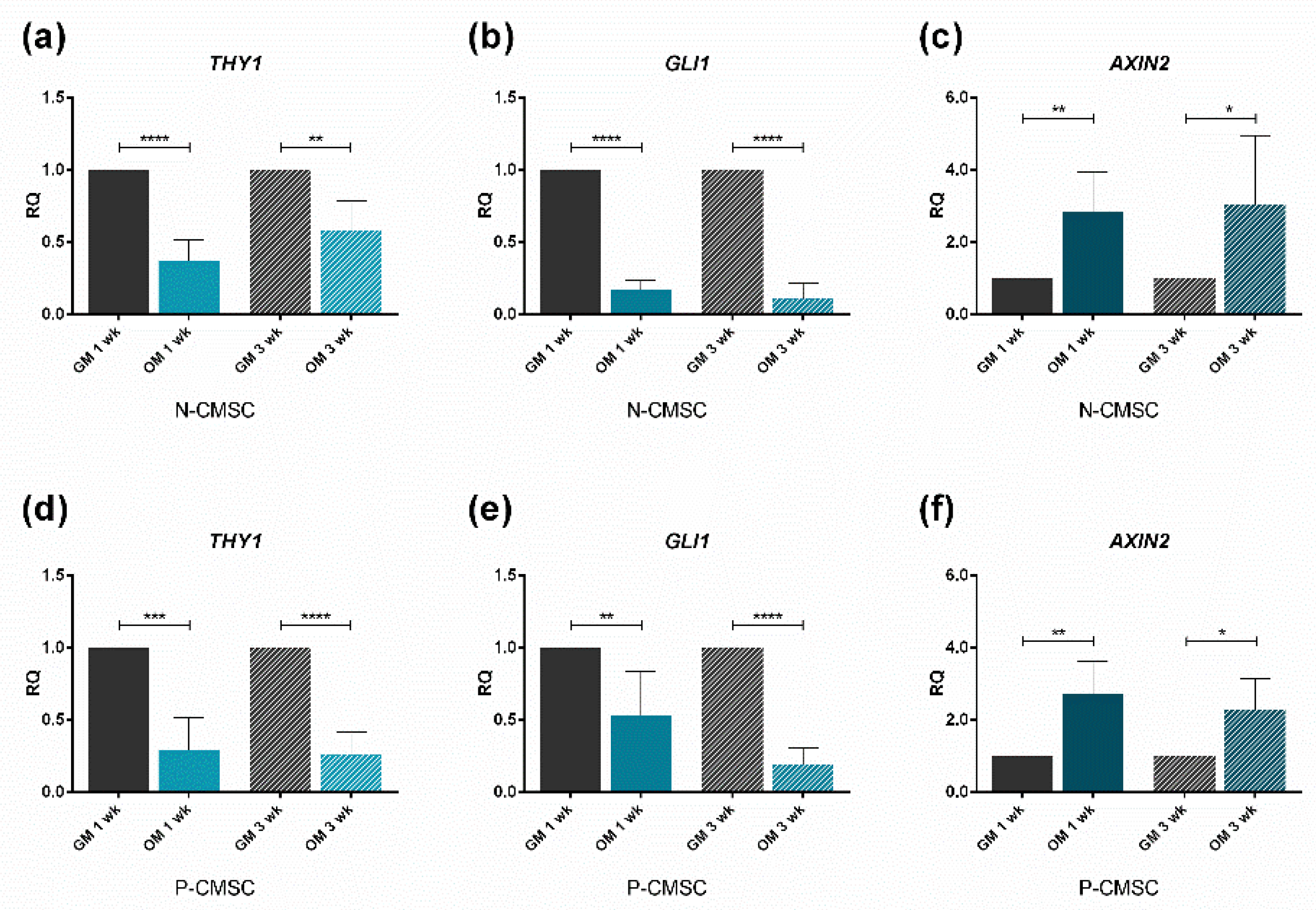
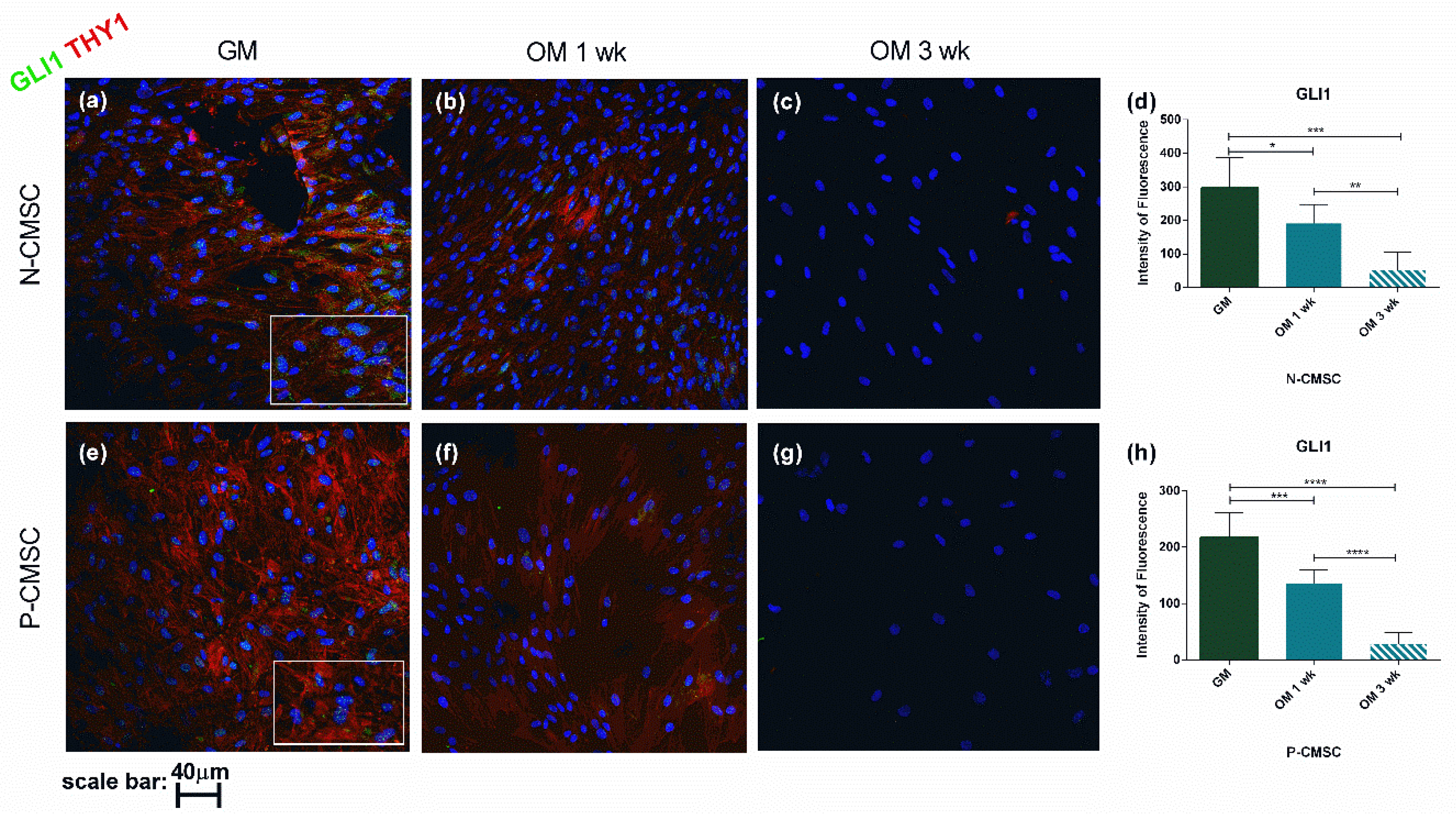
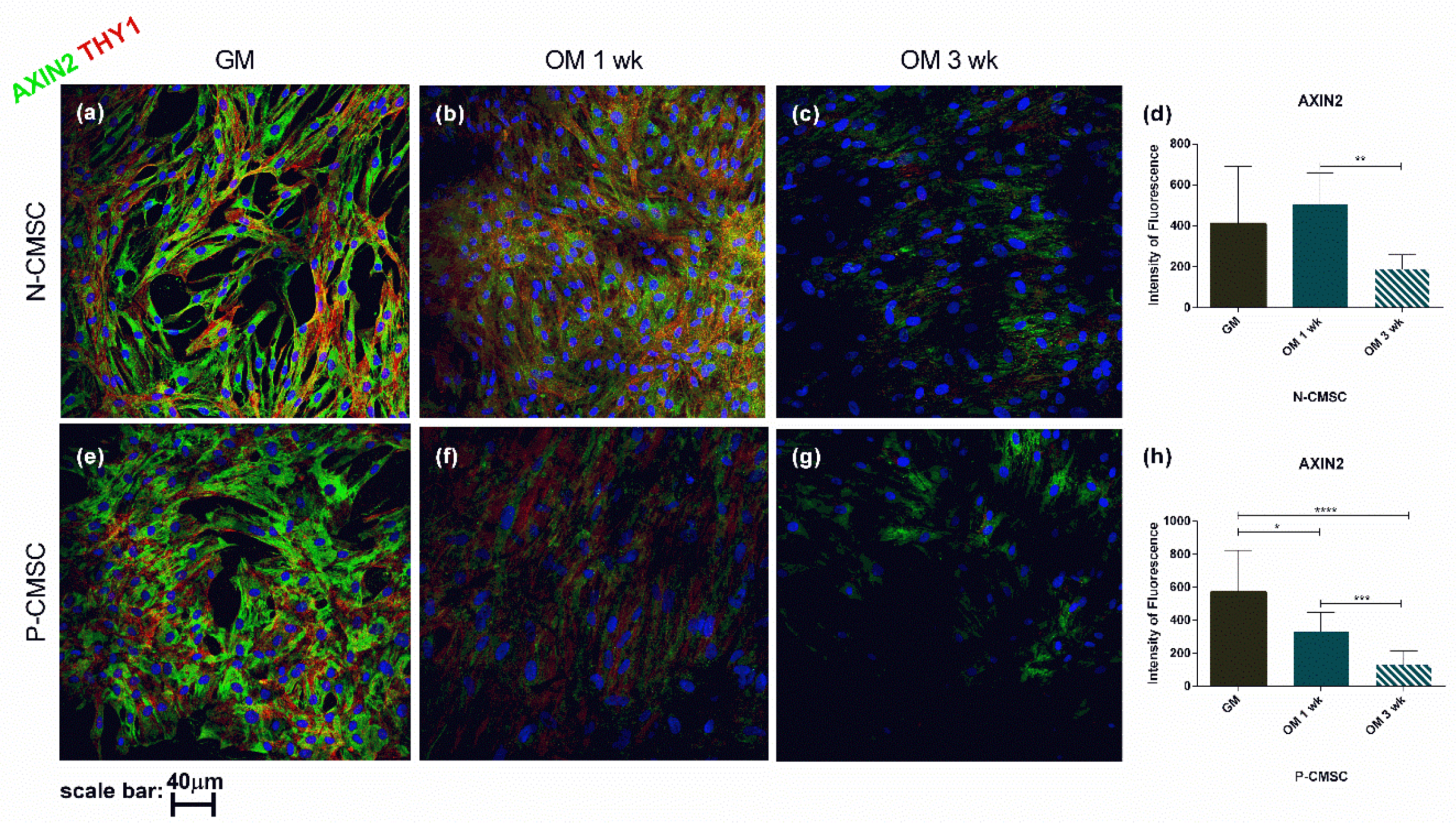
2.3. Expression of Calvarial Stem Cell Markers during Osteogenic Induction
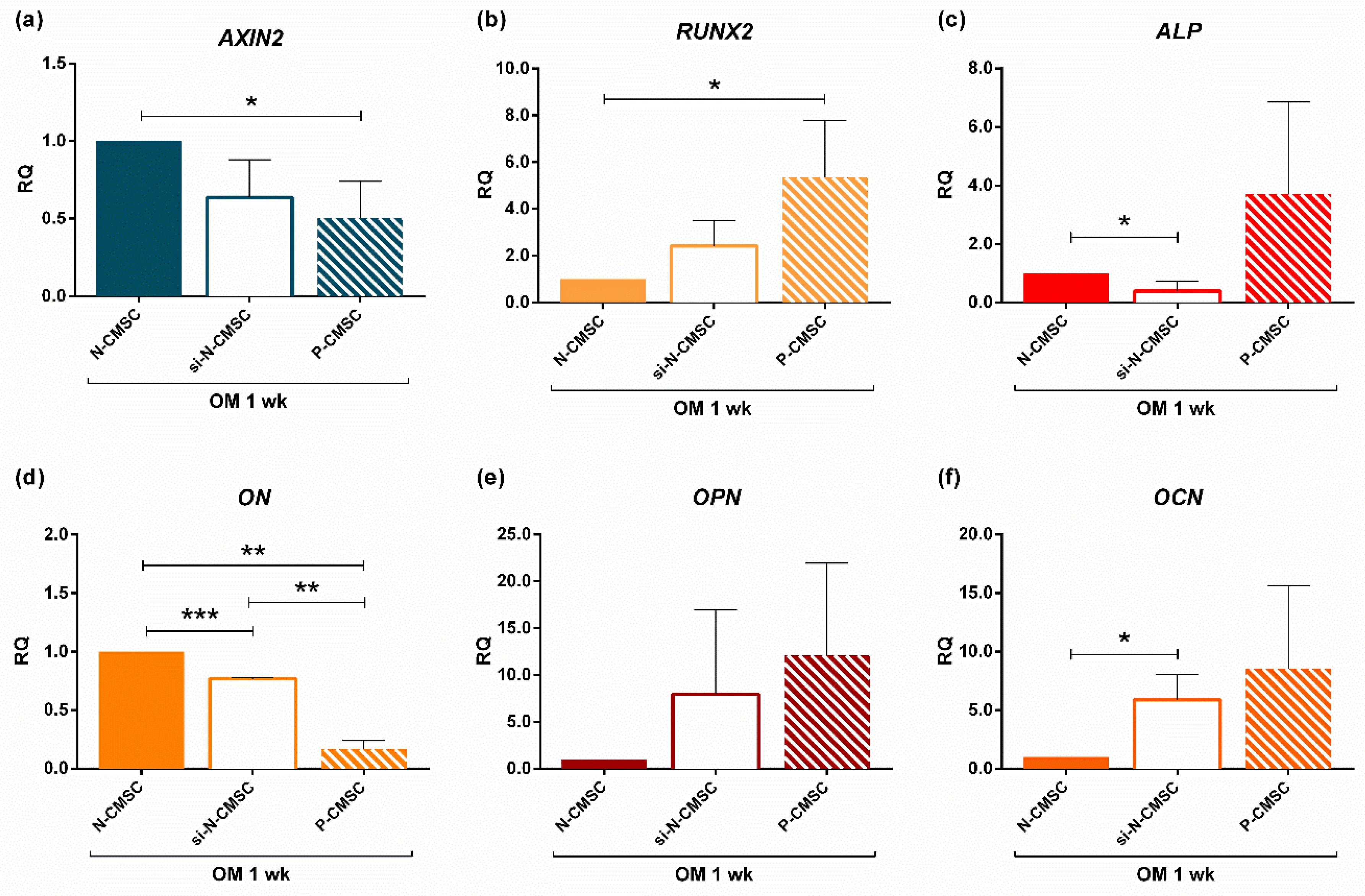
2.4. AXIN2 Involvement in the Ossification Process in NCS
3. Discussion
4. Materials and Methods
4.1. Patient Enrolment and Specimen Collection
4.2. Calvarial Mesenchymal Stromal Cells (CMSC) Isolation and Culture
4.3. Osteogenic Induction
4.4. Gene Expression Analysis
4.5. Histological Analyses
4.6. Immunofluorescence Analysis
4.7. AXIN2 Silencing in N-CMSC
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MSC | Mesenchymal stromal cells |
| WNT | Wingless-type MMTV integration site family |
| BMP | Bone morphogenetic protein family |
| FGF | Fibroblast growth factor family |
| HH | Hedgehog signalling |
| CS | Craniosynostosis |
| NCS | Nonsyndromic craniosynostosis |
| BM | Bone marrow |
| HSC | Haematopoietic stem cell |
| TEK/Tie2 | TEK receptor tyrosine kinase |
| ITGAV/AlphaV | Integrin subunit alpha V |
| GLI1 | GLI Family Zinc Finger 1 |
| THY1 | Thy-1 cell surface antigen |
| PRX1 | Paired Related Homeobox 1 |
| ENPEP | Glutamyl aminopeptidase |
| CMSC | Calvarial mesenchymal stromal cells |
| N-CMSC | Normal-CMSC |
| P-CMSC | Pathologic-CMSC |
| BM-MSC | Bone marrow mesenchymal stromal cells |
| RUNX2 | RUNX family transcription factor 2 |
| ALP | Alkaline phosphatases |
| ON | Osteonectin |
| OPN | Osteopontin |
| OCN | Osteocalcin |
| OM | Osteogenic medium |
| GM | Growth medium |
| RQ | Relative quantity |
| siRNAs | Short interfering RNAs |
| si-N-CMSC | Silenced N-CMSC |
| PTCH1 | Patched 1 |
| EDTA | Ethylenediaminetetraacetic Acid |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| FBS | Fetal bovine serum |
| qPCR | Quantitative Real Time PCR |
| ACTB | Beta-actin |
| PBS | Phosphate buffer saline |
| RT | Room temperature |
| BSA | Bovine serum albumin |
References
- Chai, Y.; Maxson, R.E. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006, 235, 2353–2375. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.K.; Miyake, T. All for one and one for all: Condensations and the initiation of skeletal development. BioEssays 2000, 22, 138–147. [Google Scholar] [CrossRef]
- Lana-Elola, E.; Rice, R.; Grigoriadis, A.E.; Rice, D.P.C. Cell fate specification during calvarial bone and suture development. Dev. Biol. 2007, 311, 335–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilk, K.; Yeh, S.-C.A.; Mortensen, L.J.; Ghaffarigarakani, S.; Lombardo, C.M.; Bassir, S.H.; Aldawood, Z.A.; Lin, C.P.; Intini, G. Postnatal calvarial skeletal stem cells expressing PRX1 reside exclusively in the calvarial sutures and are required for bone regeneration. Stem Cell Rep. 2017, 8, 933–946. [Google Scholar] [CrossRef] [PubMed]
- Opperman, L.A. Cranial sutures as intramembranous bone growth sites. Dev. Dyn. 2000, 219, 472–485. [Google Scholar] [CrossRef]
- Lattanzi, W.; Parolisi, R.; Barba, M.; Bonfanti, L. Osteogenic and neurogenic stem cells in their own place: Unraveling differences and similarities between niches. Front. Cell. Neurosci. 2015, 9, 455. [Google Scholar] [CrossRef] [Green Version]
- Flaherty, K.; Singh, N.; Richtsmeier, J.T. Understanding craniosynostosis as a growth disorder. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 429–459. [Google Scholar] [CrossRef] [Green Version]
- Lattanzi, W.; Barba, M.; Di Pietro, L.; Boyadjiev, S.A.; Lattanz, W. Genetic advances in craniosynostosis. Am. J. Med. Genet. Part A 2017, 173, 1406–1429. [Google Scholar] [CrossRef] [Green Version]
- Twigg, S.R.F.; Wilkie, A.O.M. A genetic-pathophysiological framework for craniosynostosis. Am. J. Hum. Genet. 2015, 97, 359–377. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, T.; Mirando, A.J.; Deng, C.X.; Hsu, W. The balance of WNT and FGF signaling influences mesenchymal stem cell fate during skeletal development. Sci. Signal. 2010, 3, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Feng, J.; Ho, T.-V.; Grimes, W.; Urata, M.; Chai, Y. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 2015, 17, 386–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkie, A.O.M.; Johnson, D.; Wall, S.A. Clinical genetics of craniosynostosis. Curr. Opin. Pediatr. 2017, 29, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Lajeunie, E.; Le Merrer, M.; Bonaïti-Pellie, C.; Marchac, D.; Renier, D. Genetic study of nonsyndromic coronal craniosynostosis. Am. J. Med. Genet. 1995, 55, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Le, T.; Zhu, Y.; Elakis, G.; Turner, A.; Lo, W.; Venselaar, H.; Verrenkamp, C.A.; Snow, N.; Mowat, D.; et al. A craniosynostosis massively parallel sequencing panel study in 309 Australian and New Zealand patients: Findings and recommendations. Genet. Med. 2018, 20, 1061–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lattanzi, W.; Bukvic, N.; Barba, M.; Tamburrini, G.; Bernardini, C.; Michetti, F.; Di Rocco, C. Genetic basis of single-suture synostoses: Genes, chromosomes and clinical implications. Child’s Nerv. Syst. 2012, 28, 1301–1310. [Google Scholar] [CrossRef]
- Justice, C.M.; Cuellar, A.; Bala, K.; Sabourin, J.A.; Cunningham, M.L.; Crawford, K.; Phipps, J.M.; Zhou, Y.; Cilliers, D.; Byren, J.C.; et al. A genome-wide association study implicates the BMP7 locus as a risk factor for nonsyndromic metopic craniosynostosis. Hum. Genet. 2020. published online ahead of print. [Google Scholar] [CrossRef]
- Cuellar, A.; Bala, K.; Di Pietro, L.; Barba, M.; Yagnik, G.; Liu, J.L.; Stevens, C.; Hur, D.J.; Ingersoll, R.G.; Justice, C.M.; et al. Gain-of-function variants and overexpression of RUNX2 in patients with nonsyndromic midline craniosynostosis. Bone 2020, 115395, published online ahead of print. [Google Scholar] [CrossRef]
- Tamburrini, G.; Caldarelli, M.; Massimi, L.; Gasparini, G.; Pelo, S.; Di Rocco, C. Complex craniosynostoses: A review of the prominent clinical features and the related management strategies. Child’s Nerv. Syst. 2012, 28, 1511–1523. [Google Scholar] [CrossRef]
- Massimi, L.; Di Rocco, C. Mini-invasive surgical technique for sagittal craniosynostosis. Child’s Nerv. Syst. 2012, 28, 1341–1345. [Google Scholar] [CrossRef]
- Simpson, A.; Wong, A.L.; Bezuhly, M. Surgical correction of nonsyndromic sagittal craniosynostosis: Concepts and controversies. Ann. Plast. Surg. 2017, 78, 103–110. [Google Scholar] [CrossRef]
- Senarath-Yapa, K.; Chung, M.T.; McArdle, A.; Wong, V.W.; Quarto, N.; Longaker, M.T.; Wan, D.C. Craniosynostosis molecular pathways and future pharmacologic therapy. Organogenesis 2012, 8, 103–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Méndez-Ferrer, S.; Michurina, T.V.; Ferraro, F.; Mazloom, A.R.; MacArthur, B.D.; Lira, S.A.; Scadden, D.T.; Ma’ayan, A.; Enikolopov, G.N.; Frenette, P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 2010, 466, 829–834. [Google Scholar] [CrossRef]
- Morrison, S.J.; Scadden, D.T.; Hospital, M.G.; Biology, R.; Darwin, F. The bone marrow niche for HSCs. Nature 2015, 505, 327–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghaloo, T.L.; Chaichanasakul, T.; Bezouglaia, O.; Kang, B.; Franco, R.; Dry, S.M.; Atti, E.; Tetradis, S. Osteogenic potential of mandibular vs. Long-bone marrow stromal cells. J. Dent. Res. 2010, 89, 1293–1298. [Google Scholar] [CrossRef]
- Matsubara, T.; Suardita, K.; Ishii, M.; Sugiyama, M.; Igarashi, A.; Oda, R.; Nishimura, M.; Saito, M.; Nakagawa, K.; Yamanaka, K.; et al. Alveolar bone marrow as a cell source for regenerative medicine: Differences between alveolar and iliac bone marrow stromal cells. J. Bone Miner. Res. 2005, 20, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.K.F.; Lindau, P.; Jiang, W.; Chen, J.Y.; Zhang, L.F.; Chen, C.C.; Seita, J.; Sahoo, D.; Kim, J.B.; Lee, A.; et al. Clonal precursor of bone, cartilage, and hematopoietic niche stromal cells. Proc. Natl. Acad. Sci. USA 2013, 110, 12643–12648. [Google Scholar] [CrossRef] [Green Version]
- Doro, D.H.; Grigoriadis, A.E.; Liu, K.J. Calvarial suture-derived stem cells and their contribution to cranial bone repair. Front. Physiol. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Yu, H.M.I.; Jerchow, B.; Sheu, T.J.; Liu, B.; Costantini, F.; Puzas, J.E.; Birchmeier, W.; Hsu, W. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development 2005, 132, 1995–2005. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, T.; Jiang, M.; Abbott, A.; Yu, H.M.I.; Huang, Q.; Chrzanowska-Wodnicka, M.; Chen, E.I.; Hsu, W. Rap1b is an effector of Axin2 regulating crosstalk of signaling pathways during skeletal development. J. Bone Miner. Res. 2017, 32, 1816–1828. [Google Scholar] [CrossRef]
- Lu, X.; Beck, G.R.; Gilbert, L.C.; Camalier, C.E.; Bateman, N.W.; Hood, B.L.; Conrads, T.P.; Kern, M.J.; You, S.; Chen, H.; et al. Identification of the homeobox protein Prx1 (MHox, Prrx-1) as a regulator of osterix expression and mediator of tumor necrosis factor α action in osteoblast differentiation. J. Bone Miner. Res. 2011, 26, 209–219. [Google Scholar] [CrossRef] [Green Version]
- Lattanzi, W.; Barba, M.; Novegno, F.; Massimi, L.; Tesori, V.; Tamburrini, G.; Galgano, S.; Bernardini, C.; Caldarelli, M.; Michetti, F.; et al. Lim mineralization protein is involved in the premature calvarial ossification in sporadic craniosynostoses. Bone 2013, 52, 474–484. [Google Scholar] [CrossRef]
- Barba, M.; Di Pietro, L.; Massimi, L.; Geloso, M.C.; Frassanito, P.; Caldarelli, M.; Michetti, F.; Della Longa, S.; Romitti, P.A.; Di Rocco, C.; et al. BBS9 gene in nonsyndromic craniosynostosis: Role of the primary cilium in the aberrant ossification of the suture osteogenic niche. Bone 2018, 112, 58–70. [Google Scholar] [CrossRef]
- Friedenstein, A.J.; Petrakova, K.V.; Kurolesova, A.I.; Frolova, G.P. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 1968, 6, 230–247. [Google Scholar] [CrossRef]
- Tavassoli, M.; Crosby, W.H. Transplantation of marrow to extramedullary sites. Science 1968, 161, 54–56. [Google Scholar] [CrossRef]
- Mohammad, K.; Dakik, P.; Medkour, Y.; Mitrofanova, D.; Titorenko, V.I. Quiescence entry, maintenance, and exit in adult stem cells. Int. J. Mol. Sci. 2019, 20, 2158. [Google Scholar] [CrossRef] [Green Version]
- Cohen, M.; Kicheva, A.; Ribeiro, A.; Blassberg, R.; Page, K.M.; Barnes, C.P.; Briscoe, J. Ptch1 and Gli regulate Shh signalling dynamics via multiple mechanisms. Nat. Commun. 2015, 6, 6709. [Google Scholar] [CrossRef]
- Kim, W.K.; Meliton, V.; Bourquard, N.; Hahn, T.J.; Parhami, F. Hedgehog signaling and osteogenic differentiation in multipotent bone marrow stromal cells are inhibited by oxidative stress. J. Cell. Biochem. 2010, 111, 1199–1209. [Google Scholar] [CrossRef]
- Plaisant, M.; Fontaine, C.; Cousin, W.; Rochet, N.; Dani, C.; Peraldi, P. Activation of hedgehog signaling inhibits osteoblast differentiation of human mesenchymal stem cells. Stem Cells 2009, 27, 703–713. [Google Scholar] [CrossRef]
- Robbins, D.J.; Fei, D.L.; Riobo, N.A. The hedgehog signal transduction network. Sci. Signal. 2012, 5. [Google Scholar] [CrossRef] [Green Version]
- Pan, A.; Chang, L.; Nguyen, A.; James, A.W. A review of hedgehog signaling in cranial bone development. Front. Physiol. 2013, 4, 61. [Google Scholar] [CrossRef] [Green Version]
- Jho, E.; Zhang, T.; Domon, C.; Joo, C.-K.; Freund, J.-N.; Costantini, F. Wnt/-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002, 22, 1172–1183. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Yu, H.M.I.; Hsu, W. Craniosynostosis caused by Axin2 deficiency is mediated through distinct functions of β-catenin in proliferation and differentiation. Dev. Biol. 2007, 301, 298–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behr, B.; Longaker, M.T.; Quarto, N. Absence of endochondral ossification and craniosynostosis in posterior frontal cranial sutures of Axin2-/- Mice. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, E.; Mihci, E.; Guzel Nur, B.; Alper, O.M. A novel AXIN2 gene mutation in sagittal synostosis. Am. J. Med. Genet. Part A 2018, 176, 1976–1980. [Google Scholar] [CrossRef] [PubMed]
- Shevde, N.K.; Bendixen, A.C.; Maruyama, M.; Li, B.L.; Billmire, D.A. Enhanced activity of osteoblast differentiation factor (PEBP2αA2/CBFa1) in affected sutural osteoblasts from patients with nonsyndromic craniosynostosis. Cleft Palate-Craniofac. J. 2001, 38, 606–614. [Google Scholar] [CrossRef]
- Coussens, A.K.; Wilkinson, C.R.; Hughes, I.P.; Morris, C.P.; van Daal, A.; Anderson, P.J.; Powell, B.C. Unravelling the molecular control of calvarial suture fusion in children with craniosynostosis. BMC Genom. 2007, 8, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Cyprus, G.N.; Overlin, J.W.; Vega, R.A.; Ritter, A.M.; Olivares-Navarrete, R. Spatial regulation of gene expression in nonsyndromic sagittal craniosynostosis. J. Neurosurg. Pediatr. 2018, 22, 620–626. [Google Scholar] [CrossRef]
- Potter, A.B.; Rhodes, J.L.; Vega, R.A.; Ridder, T.; Shiang, R. Gene expression changes between patent and fused cranial sutures in a nonsyndromic craniosynostosis population. Eplasty 2015, 15, e12. [Google Scholar]
- Kim, S.D.; Yagnik, G.; Cunningham, M.L.; Kim, J.; Boyadjiev, S.A. MAPK/ERK signaling pathway analysis in primary osteoblasts from patients with non-syndromic sagittal craniosynostosis. Cleft Palate Craniofac. J. 2014, 51, 115–119. [Google Scholar] [CrossRef] [Green Version]
- Eppley, B.L. Increased bone formation and osteoblastic cell phenotype in premature cranial suture ossification (craniosynostosis). J. Craniofac. Surg. 1997, 8, 523–524. [Google Scholar] [CrossRef]
- Song, D.; Huang, S.; Zhang, L.; Liu, W.; Huang, B.; Feng, Y.; Liu, B.; He, T.C.; Huang, D.; Reid, R.R. Differential responsiveness to BMP9 between patent and fused suture progenitor cells from craniosynostosis patients. Plast. Reconstr. Surg. 2020, 145, 552e–562e. [Google Scholar] [CrossRef] [PubMed]
- Ratisoontorn, C.; Seto, M.L.; Broughton, K.M.; Cunningham, M.L. In vitro differentiation profile of osteoblasts derived from patients with Saethre-Chotzen syndrome. Bone 2005, 36, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Hunenko, O.; Karmacharya, J.; Ong, G.; Kirschner, R.E. Toward an understanding of nonsyndromic craniosynostosis: Altered patterns of TGF-β receptor and FGF receptor expression induced by intrauterine head constraint. Ann. Plast. Surg. 2001, 46, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Carinci, P.; Becchetti, E.; Baroni, T.; Carinci, F.; Pezzetti, F.; Stabellini, G.; Locci, P.; Scapoli, L.; Tognon, M.; Volinia, S.; et al. Extracellular matrix and growth factors in the pathogenesis of some craniofacial malformations. Eur. J. Histochem. 2007, 51 (Suppl. 1), 105–115. [Google Scholar]
- Barreto, S.; González-Vázquez, A.; Cameron, R.A.; O’Brien, F.J.; Murray, D.J. Identification of stiffness-induced signalling mechanisms in cells from patent and fused sutures associated with craniosynostosis. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Gene | Strand | Sequence |
|---|---|---|
| ACTB | FW | ACGGCATCGTCACCAACT |
| ACTB | REV | AACGGCAGAAGAGAGAACCA |
| THY1 | FW | TCTCCTCCCAGAACGTCACA |
| THY1 | REV | GATGCCCTCACACTTGACCA |
| ITGAV | FW | TGTGGCTGTCGGAGATTTCA |
| ITGAV | REV | TTCCCAAAGTCCTTGCTGCT |
| TEK | FW | TGAACACAGTGGCTGGGATG |
| TEK | REV | GTGTCAATCACGTTTGGGGC |
| ENPEP | FW | TGCCAGTGGCGAAAGAAGAG |
| ENPEP | REV | ACAGCAAAGCACACCAGGTA |
| GLI1 | FW | TGCACCGAGGGCCCACTCTT |
| GLI1 | REV | AGGGAGCTGGGTGAGGTGCG |
| AXIN2 | FW | CCTGGCTCCAGAAGATCACA |
| AXIN2 | REV | TCAAGCTCTGAGCCTTCAGC |
| RUNX2 | FW | GAACCCAGAAGGCACAGACA |
| RUNX2 | REV | GGATGAGGAATGCGCCCTAA |
| ALP | FW | CCGTGGCAACTCTATCTTTGG |
| ALP | REV | GCCATACAGGATGGCAGTGA |
| ON | FW | TGCTCCCCAGGCAAAAGAAG |
| ON | REV | AGGGCTTGCACTTGACCAAA |
| OPN | FW | TGAAACGAGTCAGCTGGATG |
| OPN | REV | GCTCTCATCATTGGCTTTCC |
| OCN | FW | GACTGTGACGAGTTGGCTGA |
| OCN | REV | AGCAGAGCGACACCCTAGAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pietro, L.; Barba, M.; Prampolini, C.; Ceccariglia, S.; Frassanito, P.; Vita, A.; Guadagni, E.; Bonvissuto, D.; Massimi, L.; Tamburrini, G.; et al. GLI1 and AXIN2 Are Distinctive Markers of Human Calvarial Mesenchymal Stromal Cells in Nonsyndromic Craniosynostosis. Int. J. Mol. Sci. 2020, 21, 4356. https://doi.org/10.3390/ijms21124356
Di Pietro L, Barba M, Prampolini C, Ceccariglia S, Frassanito P, Vita A, Guadagni E, Bonvissuto D, Massimi L, Tamburrini G, et al. GLI1 and AXIN2 Are Distinctive Markers of Human Calvarial Mesenchymal Stromal Cells in Nonsyndromic Craniosynostosis. International Journal of Molecular Sciences. 2020; 21(12):4356. https://doi.org/10.3390/ijms21124356
Chicago/Turabian StyleDi Pietro, Lorena, Marta Barba, Chiara Prampolini, Sabrina Ceccariglia, Paolo Frassanito, Alessia Vita, Enrico Guadagni, Davide Bonvissuto, Luca Massimi, Gianpiero Tamburrini, and et al. 2020. "GLI1 and AXIN2 Are Distinctive Markers of Human Calvarial Mesenchymal Stromal Cells in Nonsyndromic Craniosynostosis" International Journal of Molecular Sciences 21, no. 12: 4356. https://doi.org/10.3390/ijms21124356
APA StyleDi Pietro, L., Barba, M., Prampolini, C., Ceccariglia, S., Frassanito, P., Vita, A., Guadagni, E., Bonvissuto, D., Massimi, L., Tamburrini, G., Parolini, O., & Lattanzi, W. (2020). GLI1 and AXIN2 Are Distinctive Markers of Human Calvarial Mesenchymal Stromal Cells in Nonsyndromic Craniosynostosis. International Journal of Molecular Sciences, 21(12), 4356. https://doi.org/10.3390/ijms21124356









