Skeletal Muscle and the Effects of Ammonia Toxicity in Fish, Mammalian, and Avian Species: A Comparative Review Based on Molecular Research
Abstract
1. Introduction
2. Skeletal Muscle Fibers
3. Skeletal Muscle Growth
4. Myostatin
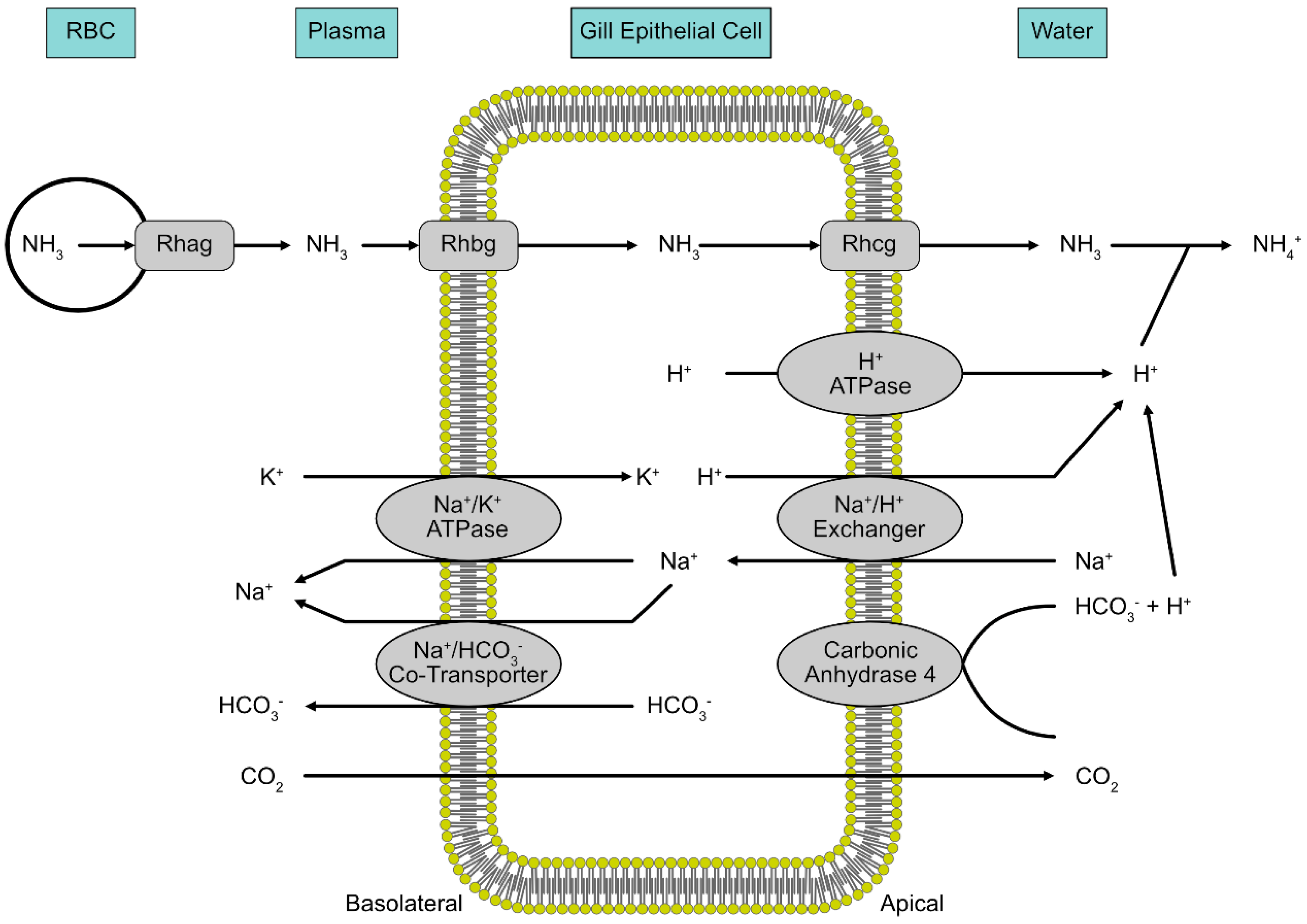
5. Ammonia
6. Ammonia Toxicity
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GDH | Glutamate dehydrogenase |
| GS | Glutamine synthetase |
| IGF-1 | Insulin-like growth factor 1 |
| MRF | Myogenic regulatory factors |
| MRF4 | Myogenic regulatory factor 4 |
| MSO | Methionine sulfoximine |
| mTOR | Mammalian rapamycin target pathway |
| Myf5 | Myogenic regulatory factor 5 |
| MyoD | Myogenic determination factor 1 |
| MSTN | Myostatin |
| PAX7 | Paired box |
| Rh | Rhesus |
| TGF | Transforming growth factor |
| UOC | Urea-ornithine cycle |
References
- Wang, W.; Walsh, W. High ammonia tolerance in fishes of the family Batrachoididae (Toadfish and Midshipmen). Aquat. Toxicol. 2000, 50, 205–219. [Google Scholar] [CrossRef]
- Wee, N.L.J.; Tng, Y.Y.M.; Cheng, H.T.; Lee, S.M.L.; Chew, S.F.; Ip, Y.K. Ammonia toxicity and tolerance in the brain of the African sharptooth catfish, Clarias gariepinus. Aquat. Toxicol. 2007, 82, 204–213. [Google Scholar] [CrossRef]
- Ip, Y.K.; Leong, M.W.F.; Sim, M.Y.; Goh, G.S.; Wong, W.P.; Chew, S.F. Chronic and acute ammonia toxicity in mudskippers, Periophthalmodon schlosseri and Boleophthalmus boddaerti: Brain ammonia and glutamine contents, and effects of methionine sulfoximine and MK801. J. Exp. Biol. 2005, 208, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Sänger, A.M.; Stoiber, W. Muscle Fiber Diversity and Plasticity. Fish Physiol. 2001, 18, 187–250. [Google Scholar]
- Greer-Walker, M. Growth and development of the Skeletal Muscle Fibres of the Cod (Gadus morhua L.). ICES J. Mar. Sci. 1970, 33, 228–244. [Google Scholar] [CrossRef]
- Zhang, G.; Swank, D.M.; Rome, L.C. Quantitative distribution of muscle fiber types in the scup Stenotomus chrysops. J. Morphol. 1996, 229, 71–81. [Google Scholar] [CrossRef]
- Rome, L.C.; Swank, D.; Corda, D. How fish power swimming. Science 1993, 261, 340–343. [Google Scholar] [CrossRef]
- Bone, Q. Locomotor muscle. In Fish Physiology; Elsevier: Amsterdam, The Netherlands, 1978. [Google Scholar]
- Jayne, B.C.; Lauder, G.V. How swimming fish use slow and fast muscle fibers: Implications for models of vertebrate muscle recruitment. J. Comp. Physiol. A 1994, 175, 123–131. [Google Scholar] [CrossRef]
- Driedzic, W.R.; Hochachka, P.W. Control of Energy Metabolism in Fish White Muscle. Am. J. Physiol. 1976, 230, 579–582. [Google Scholar] [CrossRef][Green Version]
- McKenzie, D.J. Energetics of Fish Swimming. In Encyclopedia of Fish Physiology; Elsevier: Amsterdam, The Netherlands, 2011; ISBN 9780123745453. [Google Scholar]
- Videler, J.J. Fish Swimming; Springer: Dordrecht, Netherlands, 1993; ISBN 9789401115803. [Google Scholar]
- Rowlerson, A.; Mascarello, F.; Radaelli, G.; Veggetti, A. Differentiation and growth of muscle in the fish Sparus aurata (L): II. Hyperplastic and hypertrophic growth of lateral muscle from hatching to adult. J. Muscle Res. Cell Motil. 1995, 16, 223–236. [Google Scholar] [CrossRef]
- Johnston, I.A.; Davison, W.; Goldspink, G. Energy metabolism of carp swimming muscles. J. Comp. Physiol. B 1977, 114, 203–216. [Google Scholar] [CrossRef]
- Johnston, I.A.; Patterson, S.; Ward, P.; Goldspink, G. The histochemical demonstration of myofibrillar adenosine triphosphatase activity in fish muscle. Can. J. Zool. 1974, 52, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef]
- Gerrard, D.E.; Grant, A.L. Principles of Animal Growth and Development; Kendall Hunt: Dubuque, IA, USA, 2003; ISBN 0757529860. [Google Scholar]
- Sawatari, E.; Seki, R.; Adachi, T.; Hashimoto, H.; Uji, S.; Wakamatsu, Y.; Nakata, T.; Kinoshita, M. Overexpression of the dominant-negative form of myostatin results in doubling of muscle-fiber number in transgenic medaka (Oryzias latipes). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 155, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Wiskus, K.J.; Addis, P.B.; Ma, R.-I. Distribution of βR, αR and αW Fibers in Turkey Muscles. Poult. Sci. 1976, 55, 562–572. [Google Scholar] [CrossRef]
- Pette, D.; Staron, R. Myosin Isoforms, Muscle Fiber Types, and Transitions. Microsc. Res. Tech. 2000, 50, 500–509. [Google Scholar] [CrossRef]
- Vélez, E.J.; Lutfi, E.; Azizi, S.; Perelló, M.; Salmerón, C.; Riera-Codina, M.; Ibarz, A.; Fernández-Borràs, J.; Blasco, J.; Capilla, E.; et al. Understanding fish muscle growth regulation to optimize aquaculture production. Aquaculture 2017, 467, 28–40. [Google Scholar] [CrossRef]
- Mommsen, T.P. Paradigms of Growth in Fish. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2001, 129, 207–219. [Google Scholar] [CrossRef]
- Bodine, S.C.; Stitt, T.N.; Gonzalez, M.; Kline, W.O.; Stover, G.L.; Bauerlein, R.; Zlotchenko, E.; Scrimgeour, A.; Lawrence, J.C.; Glass, D.J.; et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 2001, 3, 1014–1019. [Google Scholar] [CrossRef]
- Wigmore, P.M.; Strickland, N.C. DNA, RNA and Protein in Skeletal Muscle of Large and Small Pig Fetuses. Growth 1983, 47, 67–76. [Google Scholar] [CrossRef]
- Koumans, J.T.M.; Akster, H.A.; Booms, G.H.R.; Osse, J.W.M. Growth of carp (Cyprinus carpio) white axial muscle; hyperplasia and hypertrophy in relation to the myonucleus/sarcoplasm ratio and the occurrence of different subclasses of myogenic cells. J. Fish Biol. 1993, 43, 69–80. [Google Scholar] [CrossRef]
- Koumans, J.T.M.; Akster, H.A.; Witkam, A.; Osse, J.W.M. Numbers of muscle nuclei and myosatellite cell nuclei in red and white axial muscle during growth of the carp (Cyprinus carpio). J. Fish Biol. 1994, 44, 391–408. [Google Scholar] [CrossRef]
- Stickland, N.C. Growth and development of muscle fibres in the rainbow trout (Salmo gairdneri). J. Anat. 1983, 137, 323–333. [Google Scholar] [PubMed]
- Rowlerson, A.; Veggetti, A. Cellular Mechanisms of Post-Embryonic Muscle Growth in Aquaculture Species. Fish Physiol. 2001, 18, 103–140. [Google Scholar] [CrossRef]
- Veggetti, A.; Mascarello, F.; Scapolo, P.A.; Rowlerson, A.; Carnevali, C. Muscle growth and myosin isoform transitions during development of a small teleost fish, Poecilia reticulata (Peters) (Atheriniformes, Poeciliidae): A histochemical, immunohistochemical, ultrastructural and morphometric study. Anat. Embryol. 1993, 187, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Carpenè, E.; Veggetti, A. Increase in muscle fibres in the lateralis muscle (white portion) of Mugilidae (Pisces, Teleostei). Experientia 1981, 37, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Romanello, M.G.; Scapolo, P.A.; Luprano, S.; Mascarello, F. Post-larval growth in the lateral white muscle of the eel, Anguilla anguilla. J. Fish Biol. 1987, 30, 161–172. [Google Scholar] [CrossRef]
- Patterson, S.E.; Mook, L.B.; Devoto, S.H. Growth in the Larval Zebrafish Pectoral Fin and Trunk Musculature. Dev. Dyn. 2008, 237, 307–315. [Google Scholar] [CrossRef]
- Ahammad, A.K.S.; Asaduzzaman, M.; Asakawa, S.; Watabe, S.; Kinoshita, S. Regulation of gene expression mediating indeterminate muscle growth in teleosts. Mech. Dev. 2015, 137, 53–65. [Google Scholar] [CrossRef]
- Froehlich, J.M.; Fowler, Z.G.; Galt, N.J.; Smith, D.L.; Biga, P.R. Sarcopenia and piscines: The case for indeterminate-growing fish as unique genetic model organisms in aging and longevity research. Front. Genet. 2013, 4, 159. [Google Scholar] [CrossRef]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 is required for the specification of myogenic satellite cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef]
- Wang, Y.X.; Rudnicki, M.A. Satellite Cells, the Engines of Muscle Repair. Nat. Rev. Mol. Cell Biol. 2011, 13, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Kablar, B.; Krastel, K.; Ying, C.; Asakura, A.; Tapscott, S.J.; Rudnicki, M.A. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development 1997, 124, 4729–4738. [Google Scholar] [PubMed]
- Rudnicki, M.A.; Schnegelsberg, P.N.; Stead, R.H.; Braun, T.; Arnold, H.H.; Jaenisch, R. MyoD or Myf-5 Is Required for the Formation of Skeletal Muscle. Cell 1993, 75, 1351–1359. [Google Scholar] [CrossRef]
- Tajbakhsh, S.; Cossu, G. Establishing myogenic identity during somitogenesis. Curr. Opin. Genet. Dev. 1997, 7, 634–641. [Google Scholar] [CrossRef]
- Hasty, P.; Bradley, A.; Morris, J.H.; Edmondson, D.G.; Venuti, J.M.; Olson, E.N.; Klein, W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 1993, 364, 501–506. [Google Scholar] [CrossRef]
- Hinits, Y.; Osborn, D.P.S.; Carvajal, J.J.; Rigby, P.W.J.; Hughes, S.M. Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle. Gene Expr. Patterns 2007, 7, 738–745. [Google Scholar] [CrossRef]
- Nabeshima, Y.; Hanaoka, K.; Hayasaka, M.; Esuml, E.; Li, S.; Nonaka, I.; Nabeshima, Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 1993, 364, 532–535. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.S.; Allende, M.L.; Kelly, C.S.; Abdelhamid, A.; Murakami, T.; Andermann, P.; Doerre, O.G.; Grunwald, D.J.; Riggleman, B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development 1996, 122, 271–280. [Google Scholar]
- Cole, N.J.; Hall, T.E.; Martin, C.I.; Chapman, M.A.; Kobiyama, A.; Nihei, Y.; Watabe, S.; Johnston, I.A. Temperature and the expression of myogenic regulatory factors (MRFs) and myosin heavy chain isoforms during embryogenesis in the common carp Cyprinus carpio L. J. Exp. Biol. 2004, 207, 2111–2120. [Google Scholar] [CrossRef]
- Xie, S.Q.; Mason, P.S.; Wilkes, D.; Goldspink, G.; Fauconneau, B.; Stickland, N.C. Lower environmental temperature delays and prolongs myogenic regulatory factor expression and muscle differentiation in rainbow trout (Onchrhynchus mykiss) embryos. Differentiation 2001, 68, 106–114. [Google Scholar] [CrossRef]
- Steinbacher, P.; Haslett, J.R.; Obermayer, A.; Marschallinger, J.; Bauer, H.C.; Sänger, A.M.; Stoiber, W. MyoD and Myogenin expression during myogenic phases in brown trout: A precocious onset of mosaic hyperplasia is a prerequisite for fast somatic growth. Dev. Dyn. 2007, 236, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Barclay, R.D.; Burd, N.A.; Tyler, C.; Tillin, N.A.; Mackenzie, R.W. The Role of the IGF-1 Signaling Cascade in Muscle Protein Synthesis and Anabolic Resistance in Aging Skeletal Muscle. Front. Nutr. 2019, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, L.L.; Milasincic, D.J.; Pilch, P.F. Critical proliferation-independent window for basic fibroblast growth factor repression of myogenesis via the p42/p44 MAPK signaling pathway. J. Biol. Chem. 2001, 276, 13709–13717. [Google Scholar] [CrossRef]
- Ju, L.; Johnson, S.E. ERK2 Is Required for Efficient Terminal Differentiation of Skeletal Myoblasts. Biochem. Biophys. Res. Commun. 2006, 345, 1425–1433. [Google Scholar] [CrossRef]
- Beckman, B.R.; Larsen, D.A.; Moriyama, S.; Lee-Pawlak, B.; Dickhoff, W.W. Insulin-like Growth factor-I and Environmental Modulation of Growth During Smoltification of Spring Chinook Salmon (Oncorhynchus Tshawystscha). Gen. Comp. Endocrinol. 1998, 109, 325–335. [Google Scholar] [CrossRef]
- Pérez-Sánchez, J.; Martí-Palanca, H.; Kaushik, S.J. Ration size and protein intake affect circulating growth hormone concentration, hepatic growth hormone binding and plasma insulin-like growth factor-I immunoreactivity in a marine teleost, the gilthead sea bream (Sparus aurata). J. Nutr. 1995, 125, 546–552. [Google Scholar] [CrossRef]
- McCormick, S.D.; Kelley, K.M.; Young, G.; Nishioka, R.S.; Bern, H.A. Stimulation of coho salmon growth by insulin-like growth factor I. Gen. Comp. Endocrinol. 1992, 86, 398–406. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lawler, A.M.; Lee, S.-J. Regulation of skeletal muscle mass in mice by a new TGF-p superfamily member. Nature 1997, 387, 83–90. [Google Scholar] [CrossRef]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000, 275, 40235–40243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Topouzis, S.; Liang, L.F.; Stotish, R.L. Myostatin Signaling Through Smad2, Smad3 and Smad4 Is Regulated by the Inhibitory Smad7 by a Negative Feedback Mechanism. Cytokine 2004, 26, 262–272. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, C.; Plummer, E.; Thomas, M.; Hennebry, A.; Ashby, M.; Ling, N.; Smith, H.; Sharma, M.; Kambadur, R. Myostatin Induces Cachexia by Activating the Ubiquitin Proteolytic System Through an NF-kappaB-independent, FoxO1-dependent Mechanism. J. Cell. Physiol. 2006, 209, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Trendelenburg, A.U.; Meyer, A.; Rohner, D.; Boyle, J.; Hatakeyama, S.; Glass, D.J. Myostatin Reduces Akt/TORC1/p70S6K Signaling, Inhibiting Myoblast Differentiation and Myotube Size. Am. J. Physiol. Cell Physiol. 2009, 296, 1258–1270. [Google Scholar] [CrossRef]
- McPherron, A.C.; Lee, S.J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef] [PubMed]
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in Myostatin (GDF8) in Double-Muscled Belgian Blue and Piedmontese Cattle. Genome Res. 1997, 7, 910–915. [Google Scholar] [CrossRef]
- Acosta, J.; Carpio, Y.; Borroto, I.; González, O.; Estrada, M.P. Myostatin gene silenced by RNAi show a zebrafish giant phenotype. J. Biotechnol. 2005, 119, 324–331. [Google Scholar] [CrossRef]
- Fuentes, E.N.; Pino, K.; Navarro, C.; Delgado, I.; Valdés, J.A.; Molina, A. Transient inactivation of myostatin induces muscle hypertrophy and overcompensatory growth in zebrafish via inactivation of the SMAD signaling pathway. J. Biotechnol. 2013, 168, 295–302. [Google Scholar] [CrossRef]
- Lee, S.B.; Kim, Y.S.; Oh, M.-Y.; Jeong, I.; Seong, K.-B.; Jin, H.-J. Improving rainbow trout (Oncorhynchus mykiss) growth by treatment with a fish (Paralichthys olivaceus) myostatin prodomain expressed in soluble forms in E. coli. Aquaculture 2010, 302, 270–278. [Google Scholar] [CrossRef]
- Rescan, P.-Y.; Jutel, I.; Rallière, C. Two myostatin genes are differentially expressed in myotomal muscles of the trout (Oncorhynchus mykiss). J. Exp. Biol. 2001, 204, 3523–3529. [Google Scholar]
- Østbye, T.K.; Galloway, T.F.; Nielsen, C.; Gabestad, I.; Bardal, T.; Andersen, Ø. The two myostatin genes of Atlantic salmon (Salmo salar) are expressed in a variety of tissues. Eur. J. Biochem. 2001, 268, 5249–5257. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.B.; Goetz, F.W. Differential skeletal muscle expression of myostatin across teleost species, and the isolation of multiple myostatin isoforms. FEBS Lett. 2001, 491, 212–216. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Y.L.; Bian, W.P.; Xie, S.L.; Qi, G.L.; Liu, L.; PR, S.; JX, Z.; DS, P. Deletion of Mstna and Mstnb Impairs the Immune System and Affects Growth Performance in Zebrafish. Fish Shellfish Immunol. 2018, 72, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Maccatrozzo, L.; Bargelloni, L.; Radaelli, G.; Mascarello, F.; Patarnello, T. Characterization of the myostatin gene in the gilthead seabream (Sparus aurata): Sequence, genomic structure, and expression pattern. Mar. Biotechnol. 2001, 3, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Leggatt, R.A.; Iwama, G.K. Occurrence of polyploidy in the fishes. Rev. Fish Biol. Fish. 2003, 13, 237–246. [Google Scholar] [CrossRef]
- Gabillard, J.C.; Biga, P.R.; Rescan, P.Y.; Seiliez, I. Revisiting the Paradigm of Myostatin in Vertebrates: Insights from Fishes. Gen. Comp. Endocrinol. 2013, 194, 45–54. [Google Scholar] [CrossRef]
- Ji, S.; Losinski, R.L.; Cornelius, S.G.; Frank, G.R.; Willis, G.M.; Gerrard, D.E.; Depreux, F.F.; Spurlock, M.E. Myostatin expression in porcine tissues: Tissue specificity and developmental and postnatal regulation. Am. J. Physiol. 1998, 275, 1265–1273. [Google Scholar] [CrossRef]
- Sharma, M.; Kambadur, R.; Matthews, K.G.; Somers, W.G.; Devlin, G.P.; Conaglen, J.V.; Fowke, P.J.; Bass, J.J. Myostatin, a Transforming Growth Factor-Beta Superfamily Member, Is Expressed in Heart Muscle and Is Upregulated in Cardiomyocytes After Infarct. J. Cell. Physiol. 1999, 180, 1–9. [Google Scholar] [CrossRef]
- Rodgers, B.D.; Weber, G.M.; Sullivan, C.V.; Levine, M.A. Isolation and Characterization of Myostatin Complementary Deoxyribonucleic Acid Clones from Two Commercially Important Fish: Oreochromis mossambicusand Morone chrysops. Endocrinology 2001, 142, 1412–1418. [Google Scholar] [CrossRef][Green Version]
- Radaelli, G.; Rowlerson, A.; Mascarello, F.; Patruno, M.; Funkenstein, B. Myostatin Precursor is Present in Several Tissues in Teleost Fish: A Comparative Immunolocalization Study. Cell Tissue Res. 2003, 311, 239–250. [Google Scholar] [CrossRef]
- Carlson, C.J.; Booth, F.W.; Gordon, S.E. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am. J. Physiol. 1999, 277, 601–606. [Google Scholar] [CrossRef]
- Campbell, J.W.; Aster, P.L.; Vorhaben, J.E. Mitochondrial ammoniagenesis in liver of the channel catfish Ictalurus punctatus. Am. J. Physiol. 1983, 244, R709–R717. [Google Scholar] [CrossRef] [PubMed]
- Ip, Y.K.; Chew, S.F. Ammonia production, excretion, toxicity, and defense in fish: A review. Front. Physiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.W.; Vorhaben, J.E. Avian Mitochondrial Glutamine Metabolism. J. Biol. Chem. 1976, 251, 781–786. [Google Scholar]
- Aoki, T.T.; Brennan, M.F.; Fitzpatrick, G.F.; Knight, D.C. Leucine Meal Increases Glutamine and Total Nitrogen Release from Forearm Muscle. J. Clin. Investig. 1981, 68, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.J. 13N as a Tracer for Studying Glutamate Metabolism. Neurochem. Int. 2011, 59, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.D.; Campbell, J.W. Distribution of glutamine synthetase and carbamoyl-phosphate synthetase I in vertebrate liver. Proc. Natl. Acad. Sci. USA 1988, 85, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Randall, D.J.; Wood, C.M.; Perry, S.F.; Bergman, H.; Maloiy, G.M.; Mommsen, T.P.; Wright, P.A. Urea excretion as a strategy for survival in a fish living in a very alkaline environment. Nature 1989, 337, 165–166. [Google Scholar] [CrossRef]
- Walsh, P.J.; Danulat, E.; Mommsen, T.P. Variation in urea excretion in the gulf toadfish Opsanus beta. Mar. Biol. 1990, 106, 323–328. [Google Scholar] [CrossRef]
- Walsh, P.; Milligan, C. Effects of feeding and confinement on nitrogen metabolism and excretion in the gulf toadfish Opsanus beta. J. Exp. Biol. 1995, 198, 1559–1566. [Google Scholar]
- Ip, Y.K.; Zubaidah, R.M.; Liew, P.C.; Loong, A.M.; Hiong, K.C.; Wong, W.P.; Chew, S.F. African sharptooth catfish Clarias gariepinus does not detoxify ammonia to urea or amino acids but actively excretes ammonia during exposure to environmental ammonia. Physiol. Biochem. Zool. 2004, 77, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Blair, S.D.; Wilkie, M.P.; Edwards, S.L. Rh glycoprotein immunoreactivity in the skin and its role in extrabranchial ammonia excretion by the sea lamprey (Petromyzon marinus) in fresh water. Can. J. Zool. 2017, 95, 95–105. [Google Scholar] [CrossRef]
- Evans, D.H.; Piermarini, P.M.; Choe, K.P. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol. Rev. 2005, 85, 97–177. [Google Scholar] [CrossRef]
- Goldstein, L.; Claiborne, J.B.; Evans, D.E. Ammonia excretion by the gills of two marine teleost fish: The importance of NH4+ permeance. J. Exp. Zool. 1982, 219, 395–397. [Google Scholar] [CrossRef]
- Danulat, E.; Kempe, S. Nitrogenous waste excretion and accumulation of urea and ammonia inChalcalburnus tarichi (Cyprinidae), endemic to the extremely alkaline Lake Van (Eastern Turkey). Fish Physiol. Biochem. 1992, 9, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.A.; Wood, C.M. A New Paradigm for Ammonia Excretion in Aquatic Animals: Role of Rhesus (Rh) Glycoproteins. J. Exp. Biol. 2009, 212, 2303–2312. [Google Scholar] [CrossRef] [PubMed]
- Maetz, J. Na+/NH4+, Na+/H+ Exchanges and NH3 Movement Across the Gill of Carassius Auratus. J. Exp. Biol. 1973, 58, 255–275. [Google Scholar]
- Planelles, G. Ammonium Homeostasis and Human Rhesus Glycoproteins. Nephron. Physiol. 2007, 105, 11–17. [Google Scholar] [CrossRef]
- Nakada, T.; Westhoff, C.M.; Kato, A.; Hirose, S. Ammonia secretion from fish gill depends on a set of Rh glycoproteins. FASEB J. 2007, 21, 1067–1074. [Google Scholar] [CrossRef]
- Nawata, C.M.; Wood, C.M. mRNA expression analysis of the physiological responses to ammonia infusion in rainbow trout. J. Comp. Physiol. B 2009, 179, 799–810. [Google Scholar] [CrossRef]
- Egnew, N.; Renukdas, N.; Ramena, Y.; Yadav, A.K.; Kelly, A.M.; Lochmann, R.T.; Sinha, A.K. Physiological insights into largemouth bass (Micropterus salmoides) survival during long-term exposure to high environmental ammonia. Aquat. Toxicol. 2019, 207, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Braun, M.H.; Steele, S.L.; Perry, S.F. The responses of zebrafish (Danio rerio) to high external ammonia and urea transporter inhibition: Nitrogen excretion and expression of rhesus glycoproteins and urea transporter proteins. J. Exp. Biol. 2009, 212, 3846–3856. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Zhang, B.; Chng, Y.R.; Ong, J.L.Y.; Chew, S.F.; Wong, W.P.; Lam, S.H.; Nakada, T.; Ip, Y.K. Ammonia exposure affects the mRNA and protein expression levels of certain Rhesus glycoproteins in the gills of climbing perch. J. Exp. Biol. 2017, 220, 2916–2931. [Google Scholar] [CrossRef] [PubMed]
- Tsui, T.K.N.; Hung, C.Y.C.; Nawata, C.M.; Wilson, J.M.; Wright, P.A.; Wood, C.M. Ammonia transport in cultured gill epithelium of freshwater rainbow trout: The importance of Rhesus glycoproteins and the presence of an apical Na+/NH4+ exchange complex. J. Exp. Biol. 2009, 212, 878–892. [Google Scholar] [CrossRef]
- Iwata, K.; Deguchi, M. Metabolic Fate and Distribution of 15 N-Ammonia in an Ammonotelic Amphibious Fish, Periophthalmus modestus, Following Immersion in 15 N-Ammonium Sulfate: A Long Term Experiment. Zool. Sci. 1995, 12, 175–184. [Google Scholar] [CrossRef]
- Wright, P.A.; Steele, S.L.; Huitema, A.; Bernier, N.J. Induction of Four Glutamine Synthetase Genes in Brain of Rainbow Trout in Response to Elevated Environmental Ammonia. J. Exp. Biol. 2007, 210, 2905–2911. [Google Scholar] [CrossRef]
- Wicks, B.J.; Randall, D.J. The effect of feeding and fasting on ammonia toxicity in juvenile rainbow trout, Oncorhynchus mykiss. Aquat. Toxicol. 2002, 59, 71–82. [Google Scholar] [CrossRef]
- Banerjee, B.; Koner, D.; Bhuyan, G.; Saha, N. Differential Expression of Multiple Glutamine Synthetase Genes in Air-Breathing Magur Catfish, Clarias magur and Their Induction Under Hyper-Ammonia Stress. Gene 2018, 671, 85–95. [Google Scholar] [CrossRef]
- Veauvy, C.M.; McDonald, M.D.; Van Audekerke, J.; Vanhoutte, G.; Van Camp, N.; Van der Linden, A.; Walsh, P.J. Ammonia Affects Brain Nitrogen Metabolism but Not Hydration Status in the Gulf Toadfish (Opsanus Beta). Aquat. Toxicol. 2005, 74, 32–46. [Google Scholar] [CrossRef]
- Takahashi, H.; Koehler, R.C.; Brusilow, S.W.; Traystman, R.J. Inhibition of brain glutamine accumulation prevents cerebral edema in hyperammonemic rats. Am. J. Physiol. 1991, 261, 825–829. [Google Scholar] [CrossRef]
- Clemmesen, J.O.; Larsen, F.S.; Kondrup, J.; Hansen, B.A.; Ott, P. Cerebral herniation in patients with acute liver failure is correlated with arterial ammonia concentration. Hepatology 1999, 29, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Dasarathy, S. Consilience in Sarcopenia of Cirrhosis. J. Cachexia Sarcopenia Muscle 2012, 3, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Ganda, O.P.; Ruderman, N.B. Muscle nitrogen metabolism in chronic hepatic insufficiency. Metabolism 1976, 25, 427–435. [Google Scholar] [CrossRef]
- Stern, R.A.; Dasarathy, S.; Mozdziak, P.E. Ammonia elicits a different myogenic response in avian and murine myotubes. In Vitro Cell Dev. Biol. Anim. 2017, 53, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Dasarathy, S.; Hatzoglou, M. Hyperammonemia and Proteostasis in Cirrhosis. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Beker, A.; Vanhooser, S.L.; Swartzlander, J.H.; Teeter, R.G. Atmospheric Ammonia Concentration Effects on Broiler Growth and Performance. J. Appl. Poult. Res. 2004, 13, 5–9. [Google Scholar] [CrossRef]
- Zhang, J.; Li, C.; Tang, X.; Lu, Q.; Sa, R.; Zhang, H. High Concentrations of Atmospheric Ammonia Induce Alterations in the Hepatic Proteome of Broilers (Gallus gallus): An iTRAQ-Based Quantitative Proteomic Analysis. PLoS ONE 2015, 10, e0123596. [Google Scholar] [CrossRef]
- Smart, G.R. Investigations of the toxic mechanisms of ammonia to fish-gas exchange in rainbow trout (Salmo gairdneri) exposed to acutely lethal concentrations. J. Fish Biol. 1978, 12, 93–104. [Google Scholar] [CrossRef]
- Randall, D.J.; Tsui, T.K. Ammonia Toxicity in Fish. Mar. Pollut. Bull. 2002, 45, 17–23. [Google Scholar] [CrossRef]
- Suski, C.D.; Kieffer, J.D.; Killen, S.S.; Tufts, B.L. Sub-lethal ammonia toxicity in largemouth bass. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 381–389. [Google Scholar] [CrossRef]
- Robinette, H.R. Effect of Selected Sublethal Levels of Ammonia on the Growth of Channel Catfish (Ictalurus punctatus). Progress. Fish Cult. 1976, 38, 26–29. [Google Scholar] [CrossRef]
- Smart, G. The effect of ammonia exposure on gill structure of the rainbow trout (Salmo gairdneri). J. Fish Biol. 1976, 8, 471–475. [Google Scholar] [CrossRef]
- Daoust, P.Y.; Ferguson, H.W. The pathology of chronic ammonia toxicity in rainbow trout, Salmo gairdneri Richardson. J. Fish Dis. 1984, 7, 199–205. [Google Scholar] [CrossRef]
- Ferguson, R.I.; Ashmore, P.E.; Ashworth, P.J.; Paola, C.; Prestegaard, K.L. Measurements in a Braided River chute and lobe: 1. Flow pattern, sediment transport, and channel change. Water Resour. Res. 1992, 28, 1877–1886. [Google Scholar] [CrossRef]
- Tng, Y.Y.M.; Chew, S.F.; Wee, N.L.J.; Wong, F.K.; Wong, W.P.; Tok, C.Y.; Ip, Y.K. Acute ammonia toxicity and the protective effects of methionine sulfoximine on the swamp eel, Monopterus albus. J. Exp. Zool. A Ecol. Genet. Physiol. 2009, 311, 676–688. [Google Scholar] [CrossRef]
- Binstock, L.; Lecar, H. Ammonium Ion Currents in the Squid Giant Axon. J. Gen. Physiol. 1969, 53, 342–361. [Google Scholar] [CrossRef]
- Cooper, A.J.; Plum, F. Biochemistry and Physiology of Brain Ammonia. Physiol. Rev. 1987, 67, 440–519. [Google Scholar] [CrossRef]
- Stern, R.A.; Ashwell, C.M.; Dasarathy, S.; Mozdziak, P.E. The effect of hyperammonemia on myostatin and myogenic regulatory factor gene expression in broiler embryos. Animal 2015, 9, 992–999. [Google Scholar] [CrossRef]
- McKenzie, D.J.; Shingles, A.; Taylor, E.W. Sub-lethal plasma ammonia accumulation and the exercise performance of salmonids. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 135, 515–526. [Google Scholar] [CrossRef]
- Dosdat, A.; Person-Le Ruyet, J.; Covès, D.; Dutto, G.; Gasset, E.; Le Roux, A.; Lemarié, G. Effect of chronic exposure to ammonia on growth, food utilisation and metabolism of the European sea bass (Dicentrarchus labrax). Aquat. Living Resour. 2003, 16, 509–520. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miramontes, E.; Mozdziak, P.; N. Petitte, J.; Kulus, M.; Wieczorkiewicz, M.; Kempisty, B. Skeletal Muscle and the Effects of Ammonia Toxicity in Fish, Mammalian, and Avian Species: A Comparative Review Based on Molecular Research. Int. J. Mol. Sci. 2020, 21, 4641. https://doi.org/10.3390/ijms21134641
Miramontes E, Mozdziak P, N. Petitte J, Kulus M, Wieczorkiewicz M, Kempisty B. Skeletal Muscle and the Effects of Ammonia Toxicity in Fish, Mammalian, and Avian Species: A Comparative Review Based on Molecular Research. International Journal of Molecular Sciences. 2020; 21(13):4641. https://doi.org/10.3390/ijms21134641
Chicago/Turabian StyleMiramontes, Emily, Paul Mozdziak, James N. Petitte, Magdalena Kulus, Maria Wieczorkiewicz, and Bartosz Kempisty. 2020. "Skeletal Muscle and the Effects of Ammonia Toxicity in Fish, Mammalian, and Avian Species: A Comparative Review Based on Molecular Research" International Journal of Molecular Sciences 21, no. 13: 4641. https://doi.org/10.3390/ijms21134641
APA StyleMiramontes, E., Mozdziak, P., N. Petitte, J., Kulus, M., Wieczorkiewicz, M., & Kempisty, B. (2020). Skeletal Muscle and the Effects of Ammonia Toxicity in Fish, Mammalian, and Avian Species: A Comparative Review Based on Molecular Research. International Journal of Molecular Sciences, 21(13), 4641. https://doi.org/10.3390/ijms21134641





